Test-retest reliability
Contents
Test-retest reliability#
In this notebook, the goal is to assess the test-retest reliability of each framework. For this, we used 289 subjects from the OASIS-3 dataset. The protocol followed in the OASIS-3 dataset, implied that at least two MRI images per subject were acquired in the same day within a 60-minute period. The two images per subject were pre-processed by both frameworks and the test-retest reliability within framework was assessed.
#@title
import os
import math
import matplotlib
import numpy as np
import pandas as pd
import pingouin as pg
import seaborn as sns
import matplotlib.pyplot as plt
from collections import Counter
from sklearn.linear_model import LinearRegression
from settings import RESOURCE_DIR
from utils import get_regression_metrics, bland_altman_plot
# Tables format
pd.set_option('display.float_format', lambda x: f"{x: 0.2e}")
# Visualization format
font = {'size' : 18}
matplotlib.rc('font', **font)
sns.set_theme(style="whitegrid", font_scale=2)
# Global variables
var_compare = "software"
template_name = "a2009s"
metric_analysis = "corticalThicknessAverage"
path_csv_cortical_data = "cortical_thicknesss_test_restest.csv"
pipeline1 = "ACPC_CAT12"
pipeline2 = "FREESURFER"
color_pallete = {pipeline1: "#365162", pipeline2: "#9C5315"}
var_analyse = ["r_square", "slope", "intercept"]
/home/fmachado/anaconda3/envs/fs-cat12/lib/python3.9/site-packages/outdated/utils.py:14: OutdatedPackageWarning: The package pingouin is out of date. Your version is 0.5.1, the latest is 0.5.2.
Set the environment variable OUTDATED_IGNORE=1 to disable these warnings.
return warn(
#@title
df_names_rois = pd.read_csv(os.path.join(RESOURCE_DIR, "template", f"{template_name}_atlas_labels.csv"))
df_areas = pd.read_csv(os.path.join(RESOURCE_DIR, "template", f"{template_name}_rois_areas.csv"))
Data#
#@title
df_software_raw = pd.read_csv(os.path.join(RESOURCE_DIR, "data", path_csv_cortical_data), low_memory=False)
df_software_raw.set_index("path", inplace=True)
# Get only the template ROIs
df_software_raw = df_software_raw.loc[df_software_raw.roiName.str[1:].isin(df_names_rois.label.to_list())]
df_software_raw.head(4)
| repositoryName | software | subjectID | sessionID | run | age | gender | software.1 | roiName | corticalThicknessAverage | template | cjv_run1 | cnr_run1 | snr_total_run1 | cjv_run2 | cnr_run2 | snr_total_run2 | cjv_mean | cnr_mean | snr_total_mean | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| path | ||||||||||||||||||||
| OASIS3/sub-OAS30001/ses-d0129/anat/sub-OAS30001_ses-d0129_run-01_T1w.nii.gz | OASIS3 | ACPC_CAT12 | sub-OAS30001 | ses-d0129 | 1 | 6.55e+01 | FEMALE | ACPC_CAT12 | lG_Ins_lg_and_S_cent_ins | 2.97e+00 | a2009s | 4.01e-01 | 3.41e+00 | 1.04e+01 | 3.93e-01 | 3.42e+00 | 1.03e+01 | 3.97e-01 | 3.42e+00 | 1.03e+01 |
| OASIS3/sub-OAS30001/ses-d0129/anat/sub-OAS30001_ses-d0129_run-01_T1w.nii.gz | OASIS3 | ACPC_CAT12 | sub-OAS30001 | ses-d0129 | 1 | 6.55e+01 | FEMALE | ACPC_CAT12 | lG_and_S_cingul-Ant | 2.49e+00 | a2009s | 4.01e-01 | 3.41e+00 | 1.04e+01 | 3.93e-01 | 3.42e+00 | 1.03e+01 | 3.97e-01 | 3.42e+00 | 1.03e+01 |
| OASIS3/sub-OAS30001/ses-d0129/anat/sub-OAS30001_ses-d0129_run-01_T1w.nii.gz | OASIS3 | ACPC_CAT12 | sub-OAS30001 | ses-d0129 | 1 | 6.55e+01 | FEMALE | ACPC_CAT12 | lG_and_S_cingul-Mid-Ant | 2.57e+00 | a2009s | 4.01e-01 | 3.41e+00 | 1.04e+01 | 3.93e-01 | 3.42e+00 | 1.03e+01 | 3.97e-01 | 3.42e+00 | 1.03e+01 |
| OASIS3/sub-OAS30001/ses-d0129/anat/sub-OAS30001_ses-d0129_run-01_T1w.nii.gz | OASIS3 | ACPC_CAT12 | sub-OAS30001 | ses-d0129 | 1 | 6.55e+01 | FEMALE | ACPC_CAT12 | lG_and_S_cingul-Mid-Post | 2.39e+00 | a2009s | 4.01e-01 | 3.41e+00 | 1.04e+01 | 3.93e-01 | 3.42e+00 | 1.03e+01 | 3.97e-01 | 3.42e+00 | 1.03e+01 |
Check preprocessing problems#
Some images were only ran by one of the softwares, in the code below images with preprocessing problems are discoved, and those subjects/session/run are removed from the analysis.
#@title
roi_name = df_software_raw.loc[(~df_software_raw[metric_analysis].isna()) & (df_software_raw.template == template_name)].roiName.to_list()[1]
df_group = df_software_raw[df_software_raw["roiName"] == roi_name].copy()
df_group = df_group[df_group.template == template_name].groupby(by=['path']).apply(lambda x: list(x["software"]))
# Get the images run by only one
df_problems_running = df_group[df_group.apply(len) == 1].to_frame()
software_problems = [item for sublist in df_problems_running[0].to_list() for item in sublist]
print("\n".join([f"{number_problems} images were only processed by: {soft_name}" for soft_name, number_problems in Counter(software_problems).items()]))
1 images were only processed by: FREESURFER
Exclude the subjects only run by only one software#
#@title
df_software_raw = df_software_raw[~df_software_raw.index.isin(df_problems_running.index)]
df_software = df_software_raw.reset_index().pivot(['subjectID', 'sessionID', 'template', var_compare,'roiName'], ['run'], [metric_analysis])
df_software.head(4)
| corticalThicknessAverage | ||||||
|---|---|---|---|---|---|---|
| run | 1 | 2 | ||||
| subjectID | sessionID | template | software | roiName | ||
| sub-OAS30001 | ses-d0129 | a2009s | ACPC_CAT12 | lG_Ins_lg_and_S_cent_ins | 2.97e+00 | 2.85e+00 |
| lG_and_S_cingul-Ant | 2.49e+00 | 2.41e+00 | ||||
| lG_and_S_cingul-Mid-Ant | 2.57e+00 | 2.62e+00 | ||||
| lG_and_S_cingul-Mid-Post | 2.39e+00 | 2.43e+00 | ||||
Search subjects with at least one ROI NaN#
#@title
df_rois_nan = df_software.loc[df_software[[('corticalThicknessAverage', 1), ('corticalThicknessAverage', 2)]].isna().sum(axis=1)>0]
df_rois_nan
| corticalThicknessAverage | ||||||
|---|---|---|---|---|---|---|
| run | 1 | 2 | ||||
| subjectID | sessionID | template | software | roiName | ||
| sub-OAS30896 | ses-d0439 | a2009s | FREESURFER | lS_interm_prim-Jensen | 2.13e+00 | NaN |
| sub-OAS30986 | ses-d1188 | a2009s | FREESURFER | rLat_Fis-ant-Vertical | NaN | 2.27e+00 |
| sub-OAS31073 | ses-d0196 | a2009s | ACPC_CAT12 | lG_Ins_lg_and_S_cent_ins | 3.09e+00 | NaN |
| lG_and_S_cingul-Ant | 2.35e+00 | NaN | ||||
| lG_and_S_cingul-Mid-Ant | 2.51e+00 | NaN | ||||
| ... | ... | ... | ... | |||
| FREESURFER | rS_suborbital | 1.88e+00 | NaN | |||
| rS_subparietal | 2.33e+00 | NaN | ||||
| rS_temporal_inf | 2.40e+00 | NaN | ||||
| rS_temporal_sup | 2.45e+00 | NaN | ||||
| rS_temporal_transverse | 2.26e+00 | NaN | ||||
298 rows × 2 columns
Exclude subjects with at least one ROI NaN#
#@title
df_software_raw = df_software_raw.loc[~df_software_raw.subjectID.isin(df_rois_nan.index.get_level_values(0).to_list())]
df_software = df_software.loc[~df_software.index.get_level_values(0).isin(df_rois_nan.index.get_level_values(0).to_list())]
df_software
| corticalThicknessAverage | ||||||
|---|---|---|---|---|---|---|
| run | 1 | 2 | ||||
| subjectID | sessionID | template | software | roiName | ||
| sub-OAS30001 | ses-d0129 | a2009s | ACPC_CAT12 | lG_Ins_lg_and_S_cent_ins | 2.97e+00 | 2.85e+00 |
| lG_and_S_cingul-Ant | 2.49e+00 | 2.41e+00 | ||||
| lG_and_S_cingul-Mid-Ant | 2.57e+00 | 2.62e+00 | ||||
| lG_and_S_cingul-Mid-Post | 2.39e+00 | 2.43e+00 | ||||
| lG_and_S_frontomargin | 2.11e+00 | 2.09e+00 | ||||
| ... | ... | ... | ... | ... | ... | ... |
| sub-OAS31172 | ses-d0407 | a2009s | FREESURFER | rS_suborbital | 2.16e+00 | 1.90e+00 |
| rS_subparietal | 2.09e+00 | 2.03e+00 | ||||
| rS_temporal_inf | 2.31e+00 | 2.29e+00 | ||||
| rS_temporal_sup | 2.32e+00 | 2.32e+00 | ||||
| rS_temporal_transverse | 1.80e+00 | 2.28e+00 | ||||
87616 rows × 2 columns
Demographics#
#@title
df_subjects = df_software_raw[["repositoryName", "subjectID", "sessionID", "run", "age", "snr_total_mean", "gender"]].drop_duplicates().copy()
groupby_col = ["repositoryName"]
df_summary = df_subjects.groupby(groupby_col).apply(lambda x: pd.Series({"Total of participants": len(x),
"Number males": (x["gender"]=="MALE").sum(),
"Mean and standard deviation [years]": f"{x['age'].mean(): 0.2f}+/-{x['age'].std(): 0.2f}",
"Min Age [years]": f"{x['age'].min(): 0.2f}",
"Max Age [years]": f"{x['age'].max(): 0.2f}"}))
df_ct = df_software.reset_index()[["subjectID", var_compare, "roiName", "corticalThicknessAverage"]].melt(id_vars=["subjectID", var_compare, "roiName"])
df_summary
| Total of participants | Number males | Mean and standard deviation [years] | Min Age [years] | Max Age [years] | |
|---|---|---|---|---|---|
| repositoryName | |||||
| OASIS3 | 592 | 222 | 68.94+/- 8.85 | 45.78 | 88.86 |
Analysis#
Overall Analysis#
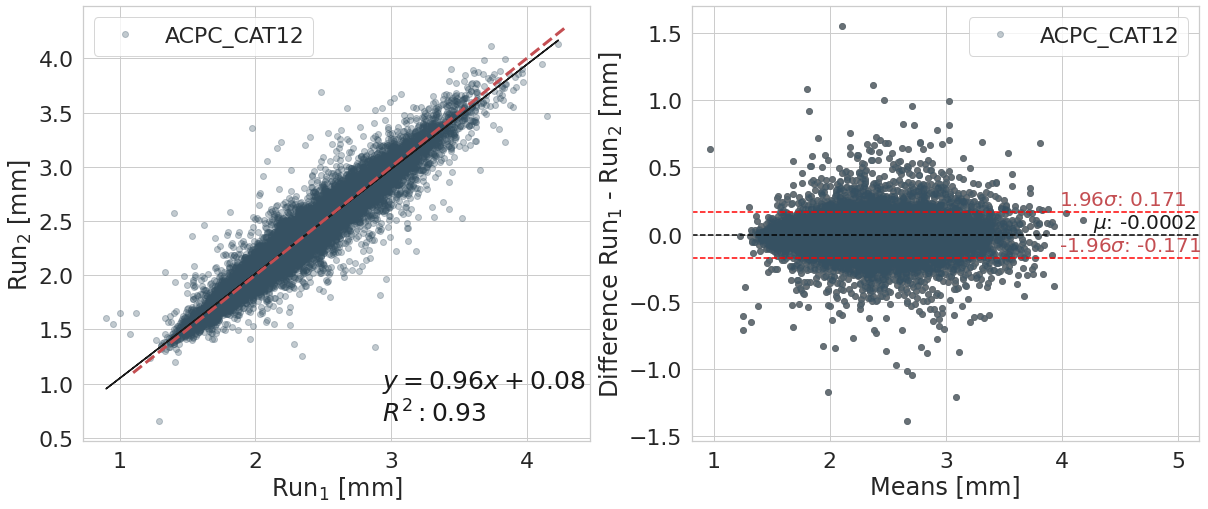
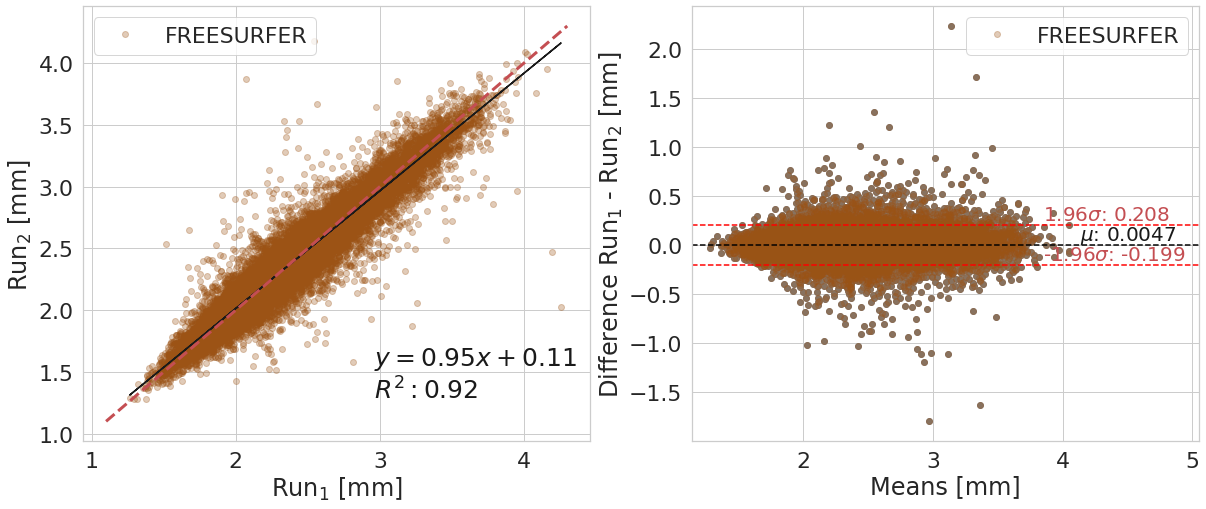
Bland-Altman plot#
#@title
y_label_txt = "Difference Run$_1$ - Run$_2$ [mm]"
for var in df_software.index.get_level_values(3).unique():
df_filt = df_software.loc[df_software.index.get_level_values(3) == var]
bland_altman_plot(df_filt.copy().reset_index(),
('corticalThicknessAverage', 1),
('corticalThicknessAverage', 2),
var_compare, color_pallete, x_label_reg='Run$_1$ [mm]', y_label_reg='Run$_2$ [mm]',
y_label_diff=y_label_txt)
from IPython.display import display, HTML
df_means = []
for soft in ['ACPC_CAT12', 'FREESURFER']:
df_soft_filter = df_software.loc[df_software.index.get_level_values(3) == soft]
diff = df_soft_filter[("corticalThicknessAverage", 1)] - df_soft_filter[("corticalThicknessAverage", 2)]
df_means_diff = df_soft_filter[[("corticalThicknessAverage", 1), ("corticalThicknessAverage", 2)]].mean().to_frame().T.rename(columns={"corticalThicknessAverage": "Cortical Thickness Average Mean"})
df_means_diff["Difference Run1 - Run 2"] = diff.mean()
test = pg.ttest(diff, 0)
display(HTML(f"<br><h5>{soft}</h5>"))
display(HTML(df_means_diff.to_html()))
display(HTML(f"<br><p>One sample t-test to verify whether the means difference is equal to zero</p>"))
display(HTML(test.to_html()))
print(f"Confidence interval (95%) of the means difference: [{round(diff.mean() - 1.96*diff.std(), 3)};{round(diff.mean() + 1.96*diff.std(), 3)}]")
ACPC_CAT12
| Cortical Thickness Average Mean | Difference Run1 - Run 2 | ||
|---|---|---|---|
| run | 1 | 2 | |
| 0 | 2.36e+00 | 2.36e+00 | -1.76e-04 |
One sample t-test to verify whether the means difference is equal to zero
| T | dof | alternative | p-val | CI95% | cohen-d | BF10 | power | |
|---|---|---|---|---|---|---|---|---|
| T-test | -4.23e-01 | 43807 | two-sided | 6.72e-01 | [-0.0, 0.0] | 2.02e-03 | 0.006 | 7.07e-02 |
Confidence interval (95%) of the means difference: [-0.171;0.171]
FREESURFER
| Cortical Thickness Average Mean | Difference Run1 - Run 2 | ||
|---|---|---|---|
| run | 1 | 2 | |
| 0 | 2.38e+00 | 2.38e+00 | 4.66e-03 |
One sample t-test to verify whether the means difference is equal to zero
| T | dof | alternative | p-val | CI95% | cohen-d | BF10 | power | |
|---|---|---|---|---|---|---|---|---|
| T-test | 9.39e+00 | 43807 | two-sided | 6.23e-21 | [0.0, 0.01] | 4.49e-02 | 7.257e+16 | 1.00e+00 |
Confidence interval (95%) of the means difference: [-0.199;0.208]
Participant Analysis#
#@title
print(f'Number of subjects in this analysis is: {len(df_software.reset_index()["subjectID"].unique())}')
Number of subjects in this analysis is: 296
Compute metrics#
#@title
all_subjects_icc = []
for var in df_ct[var_compare].unique():
for subject in df_ct["subjectID"].unique():
df_icc_roi = pg.intraclass_corr(data=df_ct.loc[(df_ct["subjectID"] == subject) & (df_ct[var_compare] == var)],
targets='roiName', raters='run',
ratings='value')
df_icc_roi["subjectID"] = subject
df_icc_roi[var_compare] = var
all_subjects_icc.append(df_icc_roi.loc[df_icc_roi["Type"]=="ICC3"])
df_icc_sub = pd.concat(all_subjects_icc)
sub_grouped_by, sub_grouped_by_stats = get_regression_metrics(df_software, ['subjectID', 'sessionID', var_compare], (metric_analysis, 1),
(metric_analysis, 2))
sub_grouped_by_stats = sub_grouped_by_stats.reset_index().set_index(["subjectID", "software"]).join(df_icc_sub.set_index(["subjectID", "software"])[["ICC"]])
sub_grouped_by_stats = sub_grouped_by_stats.reset_index().set_index(["subjectID", "sessionID", var_compare])
df_subject_info = df_subjects.loc[df_subjects["run"] ==1].reset_index().set_index(['subjectID', 'sessionID']).join(sub_grouped_by_stats)
df_subject_info = df_subject_info[~df_subject_info["slope"].isna()]
df_mean_runs = df_software.mean(axis=1).reset_index().rename(columns={0: "corticalThicknessAverage"})
df_ct_mean_runs = df_mean_runs.reset_index()[["subjectID", var_compare, "roiName", "corticalThicknessAverage"]].melt(id_vars=["subjectID", var_compare, "roiName"])
all_subjects_icc_mean_runs = []
for subject in df_ct["subjectID"].unique():
df_icc_roi_mean_runs = pg.intraclass_corr(data=df_ct_mean_runs.loc[df_ct_mean_runs["subjectID"] == subject],
targets='roiName', raters='software',
ratings='value')
df_icc_roi_mean_runs["subjectID"] = subject
df_icc_roi_mean_runs[var_compare] = var
all_subjects_icc_mean_runs.append(df_icc_roi_mean_runs.loc[df_icc_roi_mean_runs["Type"]=="ICC3"])
df_icc_sub_mean_runs = pd.concat(all_subjects_icc_mean_runs)
df_ct_means = df_mean_runs.reset_index().pivot_table(values="corticalThicknessAverage", columns="software",
index=["subjectID", "sessionID", "template", "roiName"])
sub_grouped_by_mean_runs, sub_grouped_by_stats_mean_runs = get_regression_metrics(df_ct_means, ['subjectID', 'sessionID'],
"ACPC_CAT12", "FREESURFER")
sub_grouped_by_stats_mean_runs = sub_grouped_by_stats_mean_runs.reset_index().set_index(["subjectID"]).join(df_icc_sub_mean_runs.set_index(["subjectID"])[["ICC"]])
sub_grouped_by_stats_mean_runs = sub_grouped_by_stats_mean_runs.reset_index().set_index(["subjectID", "sessionID"])
sub_grouped_by_stats_mean_runs.head(10)
| slope | intercept | r_value | p_value | std_err | r_square | ICC | ||
|---|---|---|---|---|---|---|---|---|
| subjectID | sessionID | |||||||
| sub-OAS30001 | ses-d0129 | 9.34e-01 | 2.16e-01 | 8.12e-01 | 5.31e-36 | 5.55e-02 | 6.60e-01 | 8.04e-01 |
| sub-OAS30003 | ses-d0558 | 9.17e-01 | 2.64e-01 | 8.12e-01 | 5.66e-36 | 5.45e-02 | 6.59e-01 | 8.06e-01 |
| sub-OAS30004 | ses-d1101 | 8.73e-01 | 3.13e-01 | 7.88e-01 | 1.46e-32 | 5.64e-02 | 6.21e-01 | 7.84e-01 |
| sub-OAS30007 | ses-d0061 | 9.56e-01 | 7.94e-02 | 8.44e-01 | 2.78e-41 | 5.03e-02 | 7.12e-01 | 8.37e-01 |
| sub-OAS30008 | ses-d0061 | 9.82e-01 | 6.48e-02 | 8.45e-01 | 1.74e-41 | 5.15e-02 | 7.14e-01 | 8.35e-01 |
| sub-OAS30009 | ses-d0148 | 9.90e-01 | 8.28e-03 | 8.01e-01 | 2.24e-34 | 6.12e-02 | 6.42e-01 | 7.84e-01 |
| sub-OAS30014 | ses-d0196 | 8.89e-01 | 3.07e-01 | 8.40e-01 | 1.39e-40 | 4.76e-02 | 7.05e-01 | 8.39e-01 |
| sub-OAS30015 | ses-d0116 | 8.80e-01 | 2.86e-01 | 7.88e-01 | 1.63e-32 | 5.69e-02 | 6.20e-01 | 7.83e-01 |
| sub-OAS30017 | ses-d0054 | 9.85e-01 | 5.89e-02 | 8.58e-01 | 4.69e-44 | 4.88e-02 | 7.36e-01 | 8.50e-01 |
| sub-OAS30018 | ses-d0070 | 1.02e+00 | 4.25e-02 | 7.82e-01 | 8.69e-32 | 6.69e-02 | 6.12e-01 | 7.56e-01 |
Test-retest metrics analysis#
#@title
df_subject_info.groupby(by=var_compare).apply(lambda x: pd.Series([f'{x["ICC"].mean(): 0.2f}+/-{x["ICC"].std(): 0.2f}',
f'{x["r_square"].mean(): 0.2f}+/-{x["r_square"].std(): 0.2f}'],
index=['ICC', '$R^2$']))
| ICC | $R^2$ | |
|---|---|---|
| software | ||
| ACPC_CAT12 | 0.97+/- 0.04 | 0.94+/- 0.06 |
| FREESURFER | 0.96+/- 0.04 | 0.93+/- 0.07 |
Statistical analysis – Paired t-test#
df_sub_t = df_subject_info.reset_index()
for var in ["ICC", "r_square"]:
df_result = pg.ttest(df_sub_t.loc[df_sub_t["software"] == "ACPC_CAT12"][var],
df_sub_t.loc[df_sub_t["software"] == "FREESURFER"][var], paired = True)
display(HTML(f"<br><p>{var}</p>"))
display(HTML(df_result.to_html()))
ICC
| T | dof | alternative | p-val | CI95% | cohen-d | BF10 | power | |
|---|---|---|---|---|---|---|---|---|
| T-test | 3.55e+00 | 295 | two-sided | 4.48e-04 | [0.0, 0.01] | 1.33e-01 | 29.406 | 6.28e-01 |
r_square
| T | dof | alternative | p-val | CI95% | cohen-d | BF10 | power | |
|---|---|---|---|---|---|---|---|---|
| T-test | 4.14e+00 | 295 | two-sided | 4.46e-05 | [0.01, 0.01] | 1.51e-01 | 253.979 | 7.33e-01 |
def latex_float(float_str):
if "e" in float_str:
base, exponent = float_str.split("e")
return r"{0} \times 10^{{{1}}}".format(base, int(exponent))
else:
return float_str
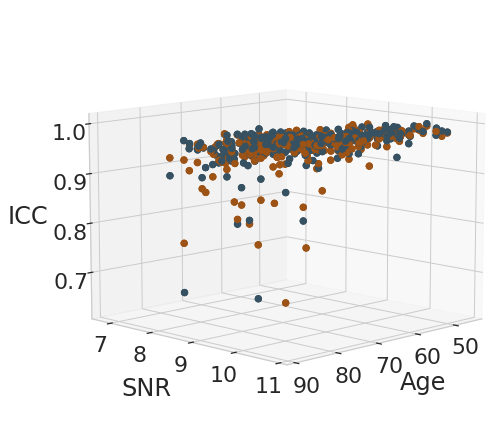
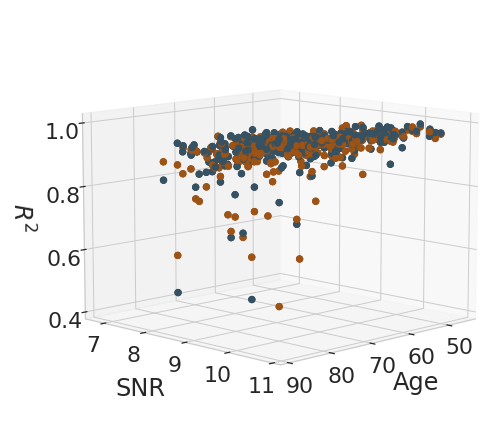
Analysis of age effect on test-retest metrics#
from IPython.display import display, Math
import re, seaborn as sns
import numpy as np
from matplotlib import pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from matplotlib.colors import ListedColormap
reliability_measures = [("ICC", "ICC"), ("r_square", "$R^2$")]
for rel_measure, label_measure in reliability_measures:
for el in df_subject_info.index.get_level_values(2).unique():
legend = el
if legend == "ACPC_CAT12":
legend = "CAT12"
data_filtered = df_subject_info.loc[df_subject_info.index.get_level_values(2) == el]
x_values = data_filtered[["age", "snr_total_mean"]]
y_values = data_filtered[rel_measure]
model = LinearRegression().fit(x_values, y_values)
age_weight = latex_float(f'{model.coef_[0]: 0.1e}')
SNR_weight = latex_float(f'{model.coef_[1]: 0.1e}')
intercept = latex_float(f'{model.intercept_: 0.1e}')
eq = f'${label_measure.replace("$", "")}_' + "{" + legend + "}" + f'={age_weight}age + {SNR_weight}SNR + {intercept}$'
display(Math(r'{}'.format(eq)))
x= df_subject_info["age"].values
y= df_subject_info["snr_total_mean"].values
z= df_subject_info[rel_measure].values
# axes instance
fig = plt.figure(figsize=(6,6))
ax = Axes3D(fig, auto_add_to_figure=False)
fig.add_axes(ax)
# get colormap from seaborn
cmap = ListedColormap(sns.color_palette("husl", 256).as_hex())
# plot
sc = ax.scatter(x, y, z, s=40, c=[color_pallete[el] for el in df_subject_info.index.get_level_values(2).values], marker='o', cmap=cmap, alpha=1)
sc = ax.scatter(x, y, z, s=40, c=[color_pallete[el] for el in df_subject_info.index.get_level_values(2).values], marker='o', cmap=cmap, alpha=1)
ax.set_xlabel('Age', labelpad=15)
ax.set_ylabel('SNR', labelpad=20)
ax.set_zlabel(label_measure, labelpad=15)
ax.view_init(elev=10., azim=45)
plt.subplots_adjust(bottom=0.2, top=2)
plt.show()
Reproducibility considering the mean of two runs#
sub_grouped_by_stats_mean_runs.apply(lambda x: f"{round(x.mean(), 2)} +/- {round(x.std(), 2)}")[["r_square", "ICC"]].to_frame()
| 0 | |
|---|---|
| r_square | 0.67 +/- 0.07 |
| ICC | 0.81 +/- 0.06 |
ROI Analysis#
Compute metrics#
#@title
roi_grouped_by, roi_grouped_by_stats = get_regression_metrics(df_software, ["roiName", var_compare],
(metric_analysis, 1),
(metric_analysis, 2))
all_rois = []
df_ct = df_software.reset_index()[["subjectID", var_compare, "roiName", "corticalThicknessAverage"]].melt(id_vars=["subjectID", var_compare, "roiName"])
for var in df_ct[var_compare].unique():
for roi_name in df_ct["roiName"].unique():
df_icc_roi = pg.intraclass_corr(data=df_ct.loc[(df_ct["roiName"] == roi_name) & (df_ct[var_compare] == var)],
targets='subjectID', raters='run',
ratings='value')
df_icc_roi["roiName"] = roi_name
df_icc_roi[var_compare] = var
all_rois.append(df_icc_roi.loc[df_icc_roi["Type"]=="ICC3"])
df_icc_rois = pd.concat(all_rois)
roi_grouped_by_stats = roi_grouped_by_stats.join(pd.concat(all_rois).set_index(["roiName", var_compare])[["ICC", "CI95%"]])
map_abbrev_to_name = df_names_rois.set_index("label")[["name"]].to_dict("series")["name"]
hem_label = {"r": "Right", "l": "Left"}
roi_grouped_by_stats["name"] = roi_grouped_by_stats.index.to_frame().roiName.apply(lambda x: map_abbrev_to_name[x[1:]])
roi_grouped_by_stats["hem"] = roi_grouped_by_stats.index.to_frame().roiName.apply(lambda x: hem_label[x[0].lower()])
roi_melted = roi_grouped_by_stats.reset_index()[[var_compare, 'r_square', 'ICC', 'CI95%', 'name', 'hem']].melt(id_vars=[var_compare, 'name', 'hem'])
df_sorted_r2_icc = pd.pivot_table(roi_melted, index="name", values=["value"], columns=[var_compare, "hem", "variable"])
dfs = []
rois_info = roi_grouped_by_stats.copy()
rois_info["label"] = roi_grouped_by_stats.index.get_level_values(0).str[1:].str.strip()
rois_info["hem"] = roi_grouped_by_stats.index.get_level_values(0).str[0]
rois_info = rois_info.reset_index().set_index(["label", "hem", var_compare])
for var in rois_info.index.get_level_values(2).unique():
for hem in rois_info.index.get_level_values(1).unique():
df_areas_metrics_var = pd.concat([df_areas, df_areas["Label"].apply(lambda x: rois_info.loc[(x, hem, var)])], axis=1)
df_areas_metrics_var[var_compare] = var
dfs.append(df_areas_metrics_var)
df_areas_metrics = pd.concat(dfs)
df_areas_metrics["hem"] = df_areas_metrics["roiName"].str[0]
df_r2 = df_areas_metrics.pivot_table(index="name", columns=["software", "hem"], values=["r_square"])
df_icc = df_areas_metrics.pivot_table(index="name", columns=["software", "hem"], values=["ICC"])
df_ci = df_areas_metrics.pivot_table(index="name", columns=["software", "hem"], values=["CI95%"])
df_ci_icc = df_icc.join(df_ci)
df_ci_icc.columns = df_ci_icc.columns.swaplevel(0, 2)
df_ci_icc.columns = df_ci_icc.columns.swaplevel(0, 1)
df_ci_icc = df_ci_icc[[df_ci_icc.columns[0], df_ci_icc.columns[-4],
df_ci_icc.columns[1], df_ci_icc.columns[-3],
df_ci_icc.columns[2], df_ci_icc.columns[-2],
df_ci_icc.columns[3], df_ci_icc.columns[-1]]]
roi_grouped_by_stats
| slope | intercept | r_value | p_value | std_err | r_square | ICC | CI95% | name | hem | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| roiName | software | ||||||||||
| lG_Ins_lg_and_S_cent_ins | ACPC_CAT12 | 7.42e-01 | 7.82e-01 | 7.70e-01 | 2.36e-59 | 3.58e-02 | 5.93e-01 | 7.70e-01 | [0.72, 0.81] | Long insular gyrus and central sulcus of the i... | Left |
| FREESURFER | 7.57e-01 | 7.19e-01 | 8.03e-01 | 3.85e-68 | 3.27e-02 | 6.45e-01 | 8.02e-01 | [0.76, 0.84] | Long insular gyrus and central sulcus of the i... | Left | |
| lG_and_S_cingul-Ant | ACPC_CAT12 | 8.40e-01 | 4.09e-01 | 9.15e-01 | 7.91e-118 | 2.16e-02 | 8.37e-01 | 9.12e-01 | [0.89, 0.93] | Anterior part of the cingulate gyrus and sulcus | Left |
| FREESURFER | 8.27e-01 | 4.42e-01 | 8.29e-01 | 3.69e-76 | 3.26e-02 | 6.87e-01 | 8.29e-01 | [0.79, 0.86] | Anterior part of the cingulate gyrus and sulcus | Left | |
| lG_and_S_cingul-Mid-Ant | ACPC_CAT12 | 9.43e-01 | 1.52e-01 | 9.59e-01 | 1.39e-163 | 1.62e-02 | 9.20e-01 | 9.59e-01 | [0.95, 0.97] | Middle-anterior part of the cingulate gyrus an... | Left |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| rS_temporal_inf | FREESURFER | 8.18e-01 | 4.29e-01 | 8.66e-01 | 2.29e-90 | 2.76e-02 | 7.49e-01 | 8.64e-01 | [0.83, 0.89] | Inferior temporal sulcus | Right |
| rS_temporal_sup | ACPC_CAT12 | 9.44e-01 | 1.38e-01 | 9.38e-01 | 4.18e-137 | 2.04e-02 | 8.79e-01 | 9.38e-01 | [0.92, 0.95] | Superior temporal sulcus | Right |
| FREESURFER | 8.84e-01 | 2.78e-01 | 9.00e-01 | 5.36e-108 | 2.50e-02 | 8.10e-01 | 9.00e-01 | [0.88, 0.92] | Superior temporal sulcus | Right | |
| rS_temporal_transverse | ACPC_CAT12 | 8.88e-01 | 2.37e-01 | 8.77e-01 | 9.65e-96 | 2.83e-02 | 7.70e-01 | 8.77e-01 | [0.85, 0.9] | Transverse temporal sulcus | Right |
| FREESURFER | 8.27e-01 | 3.83e-01 | 8.41e-01 | 1.42e-80 | 3.10e-02 | 7.08e-01 | 8.41e-01 | [0.8, 0.87] | Transverse temporal sulcus | Right |
296 rows × 10 columns
Test-retest metrics analysis#
#@title
roi_grouped_by_stats.groupby(by=var_compare).apply(lambda x: pd.Series([f'{x["ICC"].mean(): 0.2f}+/-{x["ICC"].std(): 0.2f}',
f'{x["r_square"].mean(): 0.2f}+/-{x["r_square"].std(): 0.2f}'],
index=['ICC', '$R^2$']))
| ICC | $R^2$ | |
|---|---|---|
| software | ||
| ACPC_CAT12 | 0.88+/- 0.05 | 0.78+/- 0.09 |
| FREESURFER | 0.84+/- 0.06 | 0.70+/- 0.10 |
df_roi_t = roi_grouped_by_stats.reset_index()
for var in ["r_square", "ICC"]:
df_result = pg.ttest(df_roi_t.loc[df_roi_t["software"] == "ACPC_CAT12"][var],
df_roi_t.loc[df_roi_t["software"] == "FREESURFER"][var], paired = True)
display(HTML(f"<br><h5>{var}</h5>"))
display(HTML(df_result.to_html()))
r_square
| T | dof | alternative | p-val | CI95% | cohen-d | BF10 | power | |
|---|---|---|---|---|---|---|---|---|
| T-test | 9.83e+00 | 147 | two-sided | 7.86e-18 | [0.06, 0.1] | 8.32e-01 | 6.861e+14 | 1.00e+00 |
ICC
| T | dof | alternative | p-val | CI95% | cohen-d | BF10 | power | |
|---|---|---|---|---|---|---|---|---|
| T-test | 9.28e+00 | 147 | two-sided | 2.10e-16 | [0.04, 0.06] | 8.08e-01 | 2.775e+13 | 1.00e+00 |
Mean per lobe#
#@title
pd.set_option('display.float_format', lambda x: f"{x: 0.2f}")
to_group_by = ["Area"]
df_areas_metrics_grouped = df_areas_metrics.groupby(to_group_by + [var_compare, "hem"]).mean()
df_lobes = pd.pivot_table(df_areas_metrics_grouped.reset_index(), index=to_group_by, values=["r_square"], columns=[var_compare])
df_lobes
| r_square | ||
|---|---|---|
| software | ACPC_CAT12 | FREESURFER |
| Area | ||
| Frontal Lobe | 0.76 | 0.63 |
| Insula | 0.77 | 0.72 |
| Limbic lobe | 0.77 | 0.72 |
| Parietal lobe | 0.83 | 0.73 |
| Temporal and occipital lobes | 0.80 | 0.77 |
Paired t-test#
#@title
all_df = []
for var in df_ct[var_compare].unique():
for roiname in df_ct['roiName'].unique():
df_posthoc = pg.pairwise_ttests(data=df_ct.loc[(df_ct['roiName'] == roiname) & (df_ct[var_compare] == var)],
dv='value', within='run', subject='subjectID',
parametric=True, padjust='fdr_bh', effsize='cohen')
# Pretty printing of table
df_posthoc['roiName'] = roiname
df_posthoc[var_compare] = var
all_df.append(df_posthoc)
df_f_p_value = pd.concat(all_df)
df_f_p_value.head(4)
| Contrast | A | B | Paired | Parametric | T | dof | alternative | p-unc | BF10 | cohen | roiName | software | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | run | 1 | 2 | True | True | -0.41 | 295.00 | two-sided | 0.68 | 0.071 | -0.02 | lG_Ins_lg_and_S_cent_ins | ACPC_CAT12 |
| 0 | run | 1 | 2 | True | True | -0.64 | 295.00 | two-sided | 0.52 | 0.08 | -0.02 | lG_and_S_cingul-Ant | ACPC_CAT12 |
| 0 | run | 1 | 2 | True | True | -1.21 | 295.00 | two-sided | 0.23 | 0.135 | -0.02 | lG_and_S_cingul-Mid-Ant | ACPC_CAT12 |
| 0 | run | 1 | 2 | True | True | 0.13 | 295.00 | two-sided | 0.89 | 0.066 | 0.00 | lG_and_S_cingul-Mid-Post | ACPC_CAT12 |
#@title
from scipy.stats import ttest_rel
df_f_p_value["hem"] = df_f_p_value.apply(lambda x: hem_label[x.roiName[0].lower()], axis=1)
df_f_p_value["name"] = df_f_p_value.apply(lambda x: map_abbrev_to_name[x.roiName[1:]], axis=1)
# Bonferroni correction
significance_level = .05
df_f_p_value["p-value"] = df_f_p_value["p-unc"].apply(lambda x: min(x*df_f_p_value.shape[0], 1))
df_f_p_value_rejected = df_f_p_value[df_f_p_value["p-value"] < significance_level]
df_f_p_value["cohen-p-value"] = df_f_p_value[["p-value", "cohen"]].apply(lambda x: f'{x["cohen"]: 0.1}*' if x['p-value'] < significance_level else f'{x["cohen"]: 0.1}', axis=1).astype(str)
df_p_value = pd.pivot_table(df_f_p_value.reset_index(), index="name", values=["cohen-p-value"], columns=[var_compare, "hem"], aggfunc=lambda x: ' '.join(x))
df_p_value
| cohen-p-value | ||||
|---|---|---|---|---|
| software | ACPC_CAT12 | FREESURFER | ||
| hem | Left | Right | Left | Right |
| name | ||||
| Angular gyrus | -0.01 | 0.0008 | -0.008 | -0.02 |
| Anterior occipital sulcus and preoccipital notch | -0.007 | -0.02 | 0.02 | -0.02 |
| Anterior part of the cingulate gyrus and sulcus | -0.02 | -0.05 | 0.007 | -0.01 |
| Anterior segment of the circular sulcus of the insula | -0.02 | -0.05 | 0.04 | 0.03 |
| Anterior transverse collateral sulcus | 0.06 | 0.05 | 0.07 | 0.08 |
| ... | ... | ... | ... | ... |
| Temporal pole | 0.06 | 0.04 | 0.1 | 0.1 |
| Transverse frontopolar gyri and sulci | -0.02 | 0.004 | -0.004 | 0.02 |
| Transverse temporal sulcus | -0.01 | -0.01 | 0.01 | 0.05 |
| Triangular part of the inferior frontal gyrus | -0.04 | -0.03 | 0.01 | 0.02 |
| Vertical ramus of the anterior segment of the lateral sulcus | 0.003 | 0.02 | 0.1 | 0.01 |
74 rows × 4 columns
#@title
print(f"Number of ROIs with at least on group that rejected the paired t-test hypothesis: {df_f_p_value_rejected.reset_index().roiName.unique().shape[0]}")
Number of ROIs with at least on group that rejected the paired t-test hypothesis: 1
Number of regions per site in which the hypothesis was rejected#
#@title
df_f_p_value_rejected
| Contrast | A | B | Paired | Parametric | T | dof | alternative | p-unc | BF10 | cohen | roiName | software | hem | name | p-value | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | run | 1 | 2 | True | True | 4.08 | 295.00 | two-sided | 0.00 | 196.679 | 0.14 | lG_insular_short | FREESURFER | Left | Short insular gyri | 0.02 |
Reproducibility of Cortical Thickness estimation (mean of both runs)#
df_ct_means.head(3)
| software | ACPC_CAT12 | FREESURFER | |||
|---|---|---|---|---|---|
| subjectID | sessionID | template | roiName | ||
| sub-OAS30001 | ses-d0129 | a2009s | lG_Ins_lg_and_S_cent_ins | 2.91 | 3.13 |
| lG_and_S_cingul-Ant | 2.45 | 2.59 | |||
| lG_and_S_cingul-Mid-Ant | 2.59 | 2.50 |
#@title
roi_grouped_by_mean_runs, roi_grouped_by_stats_mean_runs = get_regression_metrics(df_ct_means, ["roiName"],
("ACPC_CAT12"),
("FREESURFER"))
all_rois_mean_runs = []
df_ct_roi_means = df_mean_runs.reset_index()[["subjectID", "roiName", "corticalThicknessAverage", "software"]].melt(id_vars=["subjectID", "roiName", "software"])
for var in df_ct[var_compare].unique():
for roi_name in df_ct["roiName"].unique():
df_icc_roi_mean_runs = pg.intraclass_corr(data=df_ct_roi_means.loc[(df_ct_roi_means["roiName"] == roi_name)],
targets='subjectID', raters='software',
ratings='value')
df_icc_roi_mean_runs["roiName"] = roi_name
df_icc_roi_mean_runs[var_compare] = var
all_rois_mean_runs.append(df_icc_roi_mean_runs.loc[df_icc_roi_mean_runs["Type"]=="ICC3"])
df_icc_rois_mean_runs = pd.concat(all_rois_mean_runs)
roi_grouped_by_stats_mean_runs = roi_grouped_by_stats_mean_runs.join(pd.concat(all_rois_mean_runs).set_index(["roiName"])[["ICC", "CI95%"]])
roi_grouped_by_stats_mean_runs
| slope | intercept | r_value | p_value | std_err | r_square | ICC | CI95% | |
|---|---|---|---|---|---|---|---|---|
| roiName | ||||||||
| lG_Ins_lg_and_S_cent_ins | 0.51 | 1.51 | 0.58 | 0.00 | 0.04 | 0.33 | 0.57 | [0.49, 0.64] |
| lG_Ins_lg_and_S_cent_ins | 0.51 | 1.51 | 0.58 | 0.00 | 0.04 | 0.33 | 0.57 | [0.49, 0.64] |
| lG_and_S_cingul-Ant | 0.44 | 1.46 | 0.46 | 0.00 | 0.05 | 0.21 | 0.46 | [0.36, 0.54] |
| lG_and_S_cingul-Ant | 0.44 | 1.46 | 0.46 | 0.00 | 0.05 | 0.21 | 0.46 | [0.36, 0.54] |
| lG_and_S_cingul-Mid-Ant | 0.67 | 0.71 | 0.70 | 0.00 | 0.04 | 0.48 | 0.69 | [0.63, 0.75] |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| rS_temporal_inf | 0.48 | 1.25 | 0.51 | 0.00 | 0.05 | 0.26 | 0.51 | [0.42, 0.59] |
| rS_temporal_sup | 0.82 | 0.37 | 0.83 | 0.00 | 0.03 | 0.70 | 0.83 | [0.8, 0.87] |
| rS_temporal_sup | 0.82 | 0.37 | 0.83 | 0.00 | 0.03 | 0.70 | 0.83 | [0.8, 0.87] |
| rS_temporal_transverse | 0.77 | 0.66 | 0.60 | 0.00 | 0.06 | 0.35 | 0.58 | [0.49, 0.65] |
| rS_temporal_transverse | 0.77 | 0.66 | 0.60 | 0.00 | 0.06 | 0.35 | 0.58 | [0.49, 0.65] |
296 rows × 8 columns
pd.Series([f'{roi_grouped_by_stats_mean_runs["ICC"].mean(): 0.2f}+/-{roi_grouped_by_stats_mean_runs["ICC"].std(): 0.2f}',
f'{roi_grouped_by_stats_mean_runs["r_square"].mean(): 0.2f}+/-{roi_grouped_by_stats_mean_runs["r_square"].std(): 0.2f}'],
index=['ICC', '$R^2$']).to_frame().rename(columns={0: "mean +/- std"})
| mean +/- std | |
|---|---|
| ICC | 0.59+/- 0.21 |
| $R^2$ | 0.40+/- 0.21 |

