Reproducibility in different acquisition settings
Contents
Reproducibility in different acquisition settings#
This notebook aims to understand whether the acquisition setting and age has impact on reproducibility metrics on mature brains. In this analysis two open-data sharing initiatives were used: IXI and OASIS-3. The analysis was conducted per acquisition setting. Therefore, four groups were analysed: three groups for the IXI dataset (one per site/acquisition setting) and one group for the OASIS-3.
#@title
import os
import math
import matplotlib
import numpy as np
import pandas as pd
import pingouin as pg
import seaborn as sns
import matplotlib.pyplot as plt
from collections import Counter
from sklearn.linear_model import LinearRegression
from settings import RESOURCE_DIR
from utils import get_regression_metrics, bland_altman_plot
# Tables format
pd.set_option('display.float_format', lambda x: f"{x: 0.2e}")
# Visualization format
font = {'size' : 18}
matplotlib.rc('font', **font)
sns.set_theme(style="whitegrid", font_scale=2)
# Global variables
var_compare = "repository_and_site"
template_name = "a2009s"
metric_analysis = "corticalThicknessAverage"
path_csv_cortical_data = "cortical_thicknesss_different_acquisition_settings.csv"
color_pallete = {"IXI|Guys": "#D81B60", "IXI|HH": "#1E88E5", "IXI|IOP": "#FFC107", "OASIS3": "#004D40"}
var_analyse = ["r_square", "slope", "intercept"]
pipeline1 = "ACPC_CAT12"
pipeline2 = "FREESURFER"
/home/fmachado/anaconda3/envs/fs-cat12/lib/python3.9/site-packages/outdated/utils.py:14: OutdatedPackageWarning: The package pingouin is out of date. Your version is 0.5.1, the latest is 0.5.2.
Set the environment variable OUTDATED_IGNORE=1 to disable these warnings.
return warn(
#@title
df_names_rois = pd.read_csv(os.path.join(RESOURCE_DIR, "template", f"{template_name}_atlas_labels.csv"))
df_areas = pd.read_csv(os.path.join(RESOURCE_DIR, "template", f"{template_name}_rois_areas.csv"))
Data#
#@title
df_software_raw = pd.read_csv(os.path.join(RESOURCE_DIR, "data", path_csv_cortical_data), low_memory=False)
df_software_raw.set_index("path", inplace=True)
# Get only the template ROIs
df_software_raw = df_software_raw.loc[df_software_raw.roiName.str[1:].isin(df_names_rois.label.to_list())]
df_software_raw["repository_and_site"] = df_software_raw.apply(lambda x: x["repositoryName"] if (type(x["site"]) == float and math.isnan(x["site"])) or len(x["site"]) == 0 else x["repositoryName"]+ "|" + x["site"], axis=1)
df_software_raw.head(4)
| repositoryName | site | subjectID | sessionID | run | gender | age | software | roiName | corticalThicknessAverage | template | cjv | cnr | snr_total | repository_and_site | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| path | |||||||||||||||
| IXI/Guys/sub-002/anat/sub-002_T1w.nii.gz | IXI | Guys | sub-002 | 1 | 1 | FEMALE | 3.58e+01 | ACPC_CAT12 | lG_Ins_lg_and_S_cent_ins | 3.16e+00 | a2009s | 3.35e-01 | 4.06e+00 | 1.58e+01 | IXI|Guys |
| IXI/Guys/sub-002/anat/sub-002_T1w.nii.gz | IXI | Guys | sub-002 | 1 | 1 | FEMALE | 3.58e+01 | ACPC_CAT12 | lG_and_S_cingul-Ant | 2.56e+00 | a2009s | 3.35e-01 | 4.06e+00 | 1.58e+01 | IXI|Guys |
| IXI/Guys/sub-002/anat/sub-002_T1w.nii.gz | IXI | Guys | sub-002 | 1 | 1 | FEMALE | 3.58e+01 | ACPC_CAT12 | lG_and_S_cingul-Mid-Ant | 2.69e+00 | a2009s | 3.35e-01 | 4.06e+00 | 1.58e+01 | IXI|Guys |
| IXI/Guys/sub-002/anat/sub-002_T1w.nii.gz | IXI | Guys | sub-002 | 1 | 1 | FEMALE | 3.58e+01 | ACPC_CAT12 | lG_and_S_cingul-Mid-Post | 2.69e+00 | a2009s | 3.35e-01 | 4.06e+00 | 1.58e+01 | IXI|Guys |
Check preprocessing problems#
Some images were only ran by one of the softwares, in the code below images with preprocessing problems are discoved, and those subjects/session/run are removed from the analysis.
#@title
roi_name = df_software_raw.loc[(~df_software_raw[metric_analysis].isna()) & (df_software_raw.template == template_name)].roiName.to_list()[1]
df_group = df_software_raw[df_software_raw["roiName"] == roi_name].copy()
df_group = df_group[df_group.template == template_name].groupby(by=['path']).apply(lambda x: list(x["software"]))
# Get the images run by only one
df_problems_running = df_group[df_group.apply(len) == 1].to_frame()
software_problems = [item for sublist in df_problems_running[0].to_list() for item in sublist]
print("\n".join([f"{number_problems} images were only processed by: {soft_name}" for soft_name, number_problems in Counter(software_problems).items()]))
2 images were only processed by: FREESURFER
1 images were only processed by: ACPC_CAT12
Search subjects with at least one ROI NaN#
#@title
df_software_raw = df_software_raw[~df_software_raw.index.isin(df_problems_running.index)]
df_software = df_software_raw.reset_index().pivot(['path', 'subjectID', 'sessionID', 'run', 'template', var_compare,'roiName'], ['software'], [metric_analysis]).reset_index().set_index("path")
df_software.head(4)
| subjectID | sessionID | run | template | repository_and_site | roiName | corticalThicknessAverage | ||
|---|---|---|---|---|---|---|---|---|
| software | ACPC_CAT12 | FREESURFER | ||||||
| path | ||||||||
| IXI/Guys/sub-002/anat/sub-002_T1w.nii.gz | sub-002 | 1 | 1 | a2009s | IXI|Guys | lG_Ins_lg_and_S_cent_ins | 3.16e+00 | 3.06e+00 |
| IXI/Guys/sub-002/anat/sub-002_T1w.nii.gz | sub-002 | 1 | 1 | a2009s | IXI|Guys | lG_and_S_cingul-Ant | 2.56e+00 | 2.80e+00 |
| IXI/Guys/sub-002/anat/sub-002_T1w.nii.gz | sub-002 | 1 | 1 | a2009s | IXI|Guys | lG_and_S_cingul-Mid-Ant | 2.69e+00 | 2.63e+00 |
| IXI/Guys/sub-002/anat/sub-002_T1w.nii.gz | sub-002 | 1 | 1 | a2009s | IXI|Guys | lG_and_S_cingul-Mid-Post | 2.69e+00 | 2.58e+00 |
#@title
df_rois_nan = df_software.loc[df_software[[('corticalThicknessAverage', 'FREESURFER'), ('corticalThicknessAverage', 'ACPC_CAT12')]].isna().sum(axis=1)>0]
df_rois_nan
| subjectID | sessionID | run | template | repository_and_site | roiName | corticalThicknessAverage | ||
|---|---|---|---|---|---|---|---|---|
| software | ACPC_CAT12 | FREESURFER | ||||||
| path | ||||||||
| IXI/HH/sub-275/anat/sub-275_T1w.nii.gz | sub-275 | 1 | 1 | a2009s | IXI|HH | rLat_Fis-ant-Vertical | 3.10e+00 | NaN |
| IXI/IOP/sub-373/anat/sub-373_T1w.nii.gz | sub-373 | 1 | 1 | a2009s | IXI|IOP | lS_interm_prim-Jensen | 2.53e+00 | NaN |
| OASIS3/sub-OAS30979/ses-d0051/anat/sub-OAS30979_ses-d0051_T1w.nii.gz | sub-OAS30979 | ses-d0051 | 1 | a2009s | OASIS3 | rS_interm_prim-Jensen | 2.26e+00 | NaN |
| OASIS3/sub-OAS30986/ses-d1188/anat/sub-OAS30986_ses-d1188_run-01_T1w.nii.gz | sub-OAS30986 | ses-d1188 | 1 | a2009s | OASIS3 | rLat_Fis-ant-Vertical | 2.78e+00 | NaN |
Exclude subjects with at least one ROI NaN#
#@title
df_software_raw = df_software_raw.loc[~df_software_raw.index.isin(df_rois_nan.index.get_level_values(0).to_list())]
df_software = df_software.loc[~df_software.index.isin(df_rois_nan.index.get_level_values(0).to_list())]
Demographics#
#@title
df_subjects = df_software_raw[["repositoryName", "site", var_compare, "subjectID", "sessionID", "run", "age", "gender", "snr_total"]].drop_duplicates().copy()
df_subjects.site = df_subjects.site.apply(lambda x: "" if type(x)==float else x)
if len(df_subjects["repositoryName"].unique()) == 1:
groupby_col = ["site"]
else:
groupby_col = ["repositoryName", "site"]
df_summary = df_subjects.groupby(groupby_col).apply(lambda x: pd.Series({"Total of participants": len(x),
"Number males": (x["gender"]=="MALE").sum(),
"Mean and standard deviation [years]": f"{x['age'].mean(): 0.2f}+/-{x['age'].std(): 0.2f}",
"Min Age [years]": f"{x['age'].min(): 0.2f}",
"Max Age [years]": f"{x['age'].max(): 0.2f}"}))
df_summary
| Total of participants | Number males | Mean and standard deviation [years] | Min Age [years] | Max Age [years] | ||
|---|---|---|---|---|---|---|
| repositoryName | site | |||||
| IXI | Guys | 312 | 139 | 50.73+/- 15.98 | 20.07 | 86.20 |
| HH | 179 | 85 | 47.63+/- 16.61 | 20.17 | 81.94 | |
| IOP | 67 | 24 | 42.13+/- 16.60 | 19.98 | 86.32 | |
| OASIS3 | 491 | 199 | 68.33+/- 8.86 | 42.66 | 95.20 |
Analysis#
Overall Analysis#
Bland-Altman plot#
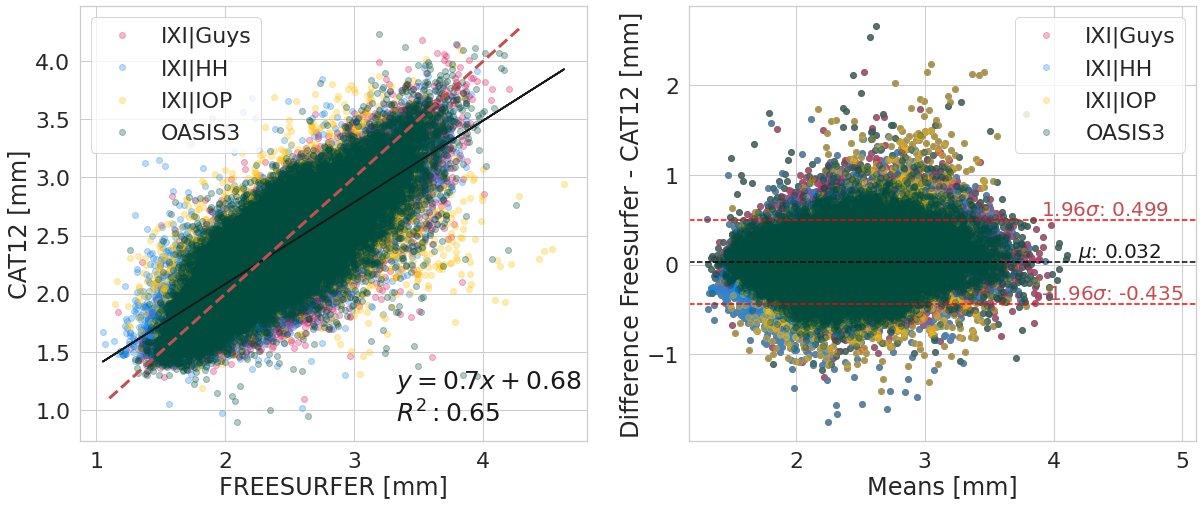
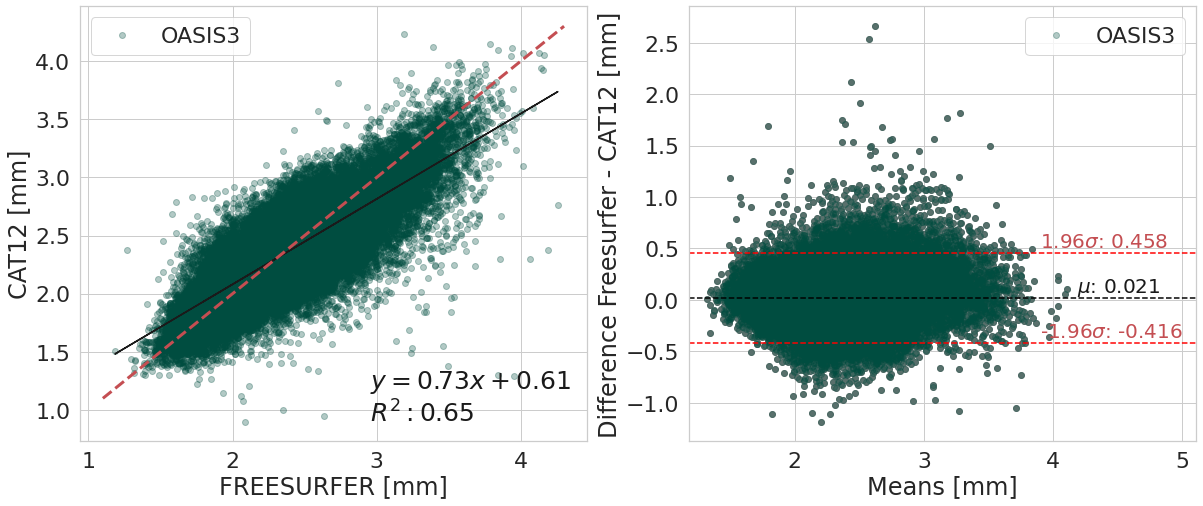
#@title
y_label_txt = "Difference Freesurfer - CAT12 [mm]"
bland_altman_plot(df_software.copy().reset_index(),
('corticalThicknessAverage', 'FREESURFER'),
('corticalThicknessAverage', 'ACPC_CAT12'),
var_compare, color_pallete, x_label_reg="FREESURFER [mm]", y_label_reg="CAT12 [mm]",
y_label_diff="Difference Freesurfer - CAT12 [mm]")
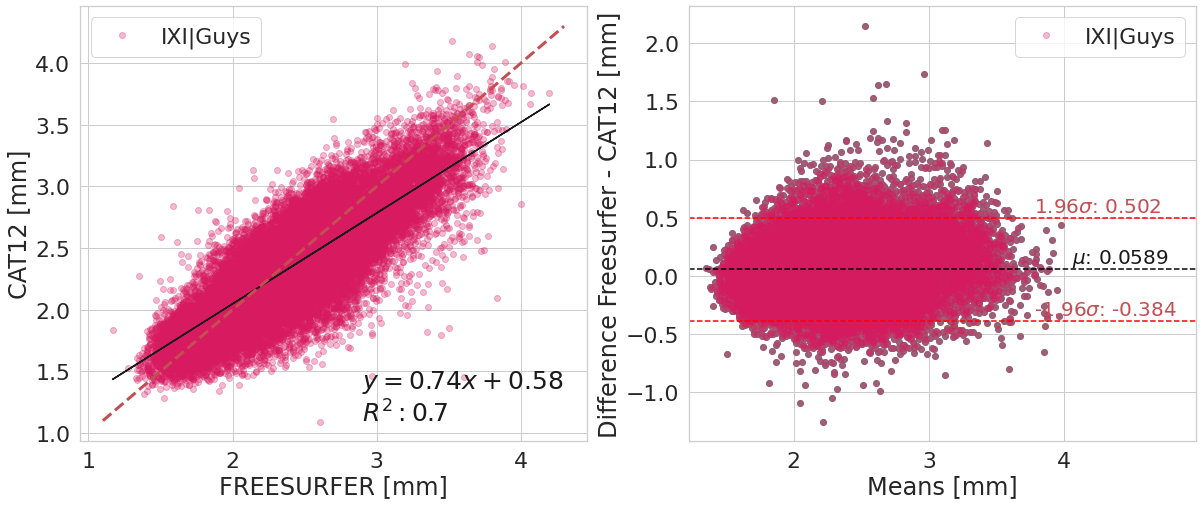
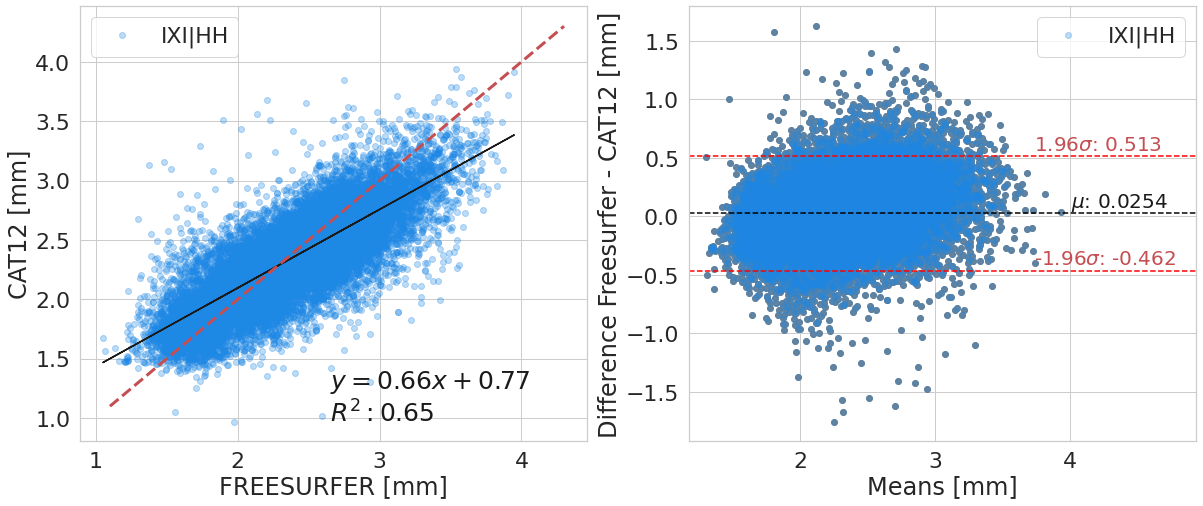
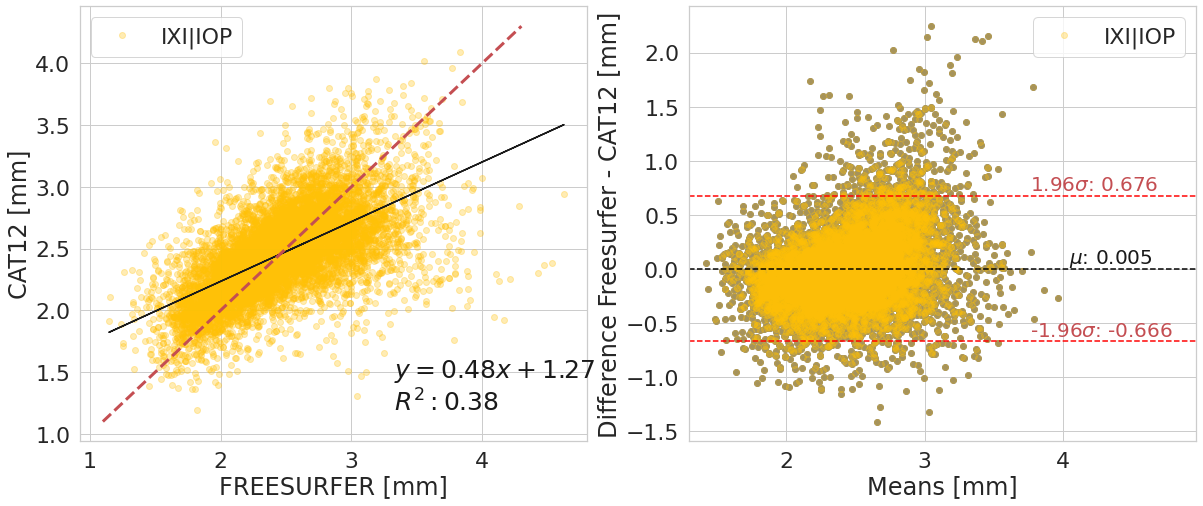
#@title
y_label_txt = "Difference Freesurfer - CAT12 [mm]"
for var_compared_value in pd.unique(df_software[var_compare]):
bland_altman_plot(df_software.loc[df_software[var_compare]==var_compared_value].copy().reset_index(),
('corticalThicknessAverage', 'FREESURFER'),
('corticalThicknessAverage', 'ACPC_CAT12'),
var_compare, color_pallete, x_label_reg="FREESURFER [mm]", y_label_reg="CAT12 [mm]",
y_label_diff="Difference Freesurfer - CAT12 [mm]")
from IPython.display import display, HTML
df_soft_filter = df_software
diff = (df_soft_filter[("corticalThicknessAverage", "FREESURFER")] - df_soft_filter[("corticalThicknessAverage", "ACPC_CAT12")])
test = pg.ttest(diff, 0)
df_means_diff = df_soft_filter[[("corticalThicknessAverage", "FREESURFER"),
("corticalThicknessAverage", "ACPC_CAT12")]].mean().to_frame().T.rename(columns={"corticalThicknessAverage": "Cortical Thickness Average Mean"})
df_means_diff["Difference FREESURFER - CAT12"] = diff.mean()
test = pg.ttest(diff, 0)
display(HTML(df_means_diff.to_html()))
display(HTML(f"<br><p>One sample t-test to verify whether the means difference is equal to zero</p>"))
display(HTML(test.to_html()))
print(f"Confidence interval (95%) of the means difference: [{round(diff.mean() - 1.96*diff.std(), 3)};{round(diff.mean() + 1.96*diff.std(), 3)}]")
/home/fmachado/anaconda3/envs/fs-cat12/lib/python3.9/site-packages/pingouin/bayesian.py:146: RuntimeWarning: divide by zero encountered in double_scalars
bf10 = 1 / ((1 + t**2 / df)**(-(df + 1) / 2) / integr)
/home/fmachado/anaconda3/envs/fs-cat12/lib/python3.9/site-packages/pingouin/bayesian.py:146: RuntimeWarning: divide by zero encountered in double_scalars
bf10 = 1 / ((1 + t**2 / df)**(-(df + 1) / 2) / integr)
| Cortical Thickness Average Mean | Difference FREESURFER - CAT12 | ||
|---|---|---|---|
| software | FREESURFER | ACPC_CAT12 | |
| 0 | 2.39e+00 | 2.36e+00 | 3.20e-02 |
One sample t-test to verify whether the means difference is equal to zero
| T | dof | alternative | p-val | CI95% | cohen-d | BF10 | power | |
|---|---|---|---|---|---|---|---|---|
| T-test | 5.29e+01 | 155251 | two-sided | 0.00e+00 | [0.03, 0.03] | 1.34e-01 | inf | 1.00e+00 |
Confidence interval (95%) of the means difference: [-0.435;0.499]
#@title
df = df_software.reset_index().set_index(["subjectID", "sessionID", "run"]).join(df_subjects.reset_index().set_index(["subjectID", "sessionID", "run"]))
df.columns = ["".join(el) for el in df.columns]
df.head(3)
/tmp/ipykernel_42420/928943096.py:3: FutureWarning: merging between different levels is deprecated and will be removed in a future version. (2 levels on the left, 1 on the right)
df = df_software.reset_index().set_index(["subjectID", "sessionID", "run"]).join(df_subjects.reset_index().set_index(["subjectID", "sessionID", "run"]))
| path | template | repository_and_site | roiName | corticalThicknessAverageACPC_CAT12 | corticalThicknessAverageFREESURFER | path | repositoryName | site | repository_and_site | age | gender | snr_total | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| subjectID | sessionID | run | |||||||||||||
| sub-002 | 1 | 1 | IXI/Guys/sub-002/anat/sub-002_T1w.nii.gz | a2009s | IXI|Guys | lG_Ins_lg_and_S_cent_ins | 3.16e+00 | 3.06e+00 | IXI/Guys/sub-002/anat/sub-002_T1w.nii.gz | IXI | Guys | IXI|Guys | 3.58e+01 | FEMALE | 1.58e+01 |
| 1 | IXI/Guys/sub-002/anat/sub-002_T1w.nii.gz | a2009s | IXI|Guys | lG_and_S_cingul-Ant | 2.56e+00 | 2.80e+00 | IXI/Guys/sub-002/anat/sub-002_T1w.nii.gz | IXI | Guys | IXI|Guys | 3.58e+01 | FEMALE | 1.58e+01 | ||
| 1 | IXI/Guys/sub-002/anat/sub-002_T1w.nii.gz | a2009s | IXI|Guys | lG_and_S_cingul-Mid-Ant | 2.69e+00 | 2.63e+00 | IXI/Guys/sub-002/anat/sub-002_T1w.nii.gz | IXI | Guys | IXI|Guys | 3.58e+01 | FEMALE | 1.58e+01 |
Participant Analysis#

#@title
print(f'Number of subjects in this analysis is: {len(df_software.reset_index()["subjectID"].unique())}')
Number of subjects in this analysis is: 1049
Compute metrics#
#@title
sub_grouped_by, sub_grouped_by_stats = get_regression_metrics(df_software, ['path'], (metric_analysis, pipeline1), (metric_analysis, pipeline2))
df_ct = df_software.reset_index()[["path", var_compare, "roiName", "corticalThicknessAverage"]].melt(id_vars=["path", var_compare, "roiName"])
all_subjects_icc = []
for path_sub in df_ct["path"].unique():
df_icc_roi = pg.intraclass_corr(data=df_ct.loc[df_ct["path"] == path_sub],
targets='roiName', raters='software',
ratings='value')
df_icc_roi["path"] = path_sub
all_subjects_icc.append(df_icc_roi.loc[df_icc_roi["Type"]=="ICC3"])
df_icc_sub = pd.concat(all_subjects_icc).set_index("path")
df_subject_info = df_subjects.join(sub_grouped_by_stats).join(df_icc_sub[["ICC"]])
df_subject_info = df_subject_info[~df_subject_info["slope"].isna()]
sub_grouped_by_stats
| slope | intercept | r_value | p_value | std_err | r_square | |
|---|---|---|---|---|---|---|
| path | ||||||
| IXI/Guys/sub-002/anat/sub-002_T1w.nii.gz | 9.76e-01 | 1.71e-01 | 8.71e-01 | 5.04e-47 | 4.54e-02 | 7.59e-01 |
| IXI/Guys/sub-016/anat/sub-016_T1w.nii.gz | 1.07e+00 | -9.83e-02 | 9.00e-01 | 1.88e-54 | 4.28e-02 | 8.10e-01 |
| IXI/Guys/sub-017/anat/sub-017_T1w.nii.gz | 1.04e+00 | 1.58e-02 | 8.27e-01 | 2.77e-38 | 5.87e-02 | 6.83e-01 |
| IXI/Guys/sub-019/anat/sub-019_T1w.nii.gz | 9.44e-01 | 1.59e-01 | 7.79e-01 | 2.11e-31 | 6.29e-02 | 6.07e-01 |
| IXI/Guys/sub-020/anat/sub-020_T1w.nii.gz | 9.85e-01 | 6.31e-02 | 8.65e-01 | 1.53e-45 | 4.73e-02 | 7.48e-01 |
| ... | ... | ... | ... | ... | ... | ... |
| OASIS3/sub-OAS31164/ses-d0069/anat/sub-OAS31164_ses-d0069_run-01_T1w.nii.gz | 8.85e-01 | 2.83e-01 | 8.30e-01 | 6.43e-39 | 4.91e-02 | 6.90e-01 |
| OASIS3/sub-OAS31165/ses-d0426/anat/sub-OAS31165_ses-d0426_run-01_T1w.nii.gz | 9.27e-01 | 2.26e-01 | 7.04e-01 | 1.91e-23 | 7.74e-02 | 4.95e-01 |
| OASIS3/sub-OAS31168/ses-d0148/anat/sub-OAS31168_ses-d0148_run-01_T1w.nii.gz | 9.60e-01 | 1.11e-01 | 7.94e-01 | 2.43e-33 | 6.09e-02 | 6.30e-01 |
| OASIS3/sub-OAS31169/ses-d0620/anat/sub-OAS31169_ses-d0620_run-01_T1w.nii.gz | 9.00e-01 | 2.07e-01 | 7.98e-01 | 6.83e-34 | 5.63e-02 | 6.36e-01 |
| OASIS3/sub-OAS31172/ses-d0407/anat/sub-OAS31172_ses-d0407_run-01_T1w.nii.gz | 9.13e-01 | 2.33e-01 | 8.37e-01 | 4.61e-40 | 4.94e-02 | 7.01e-01 |
1049 rows × 6 columns
Reproducibility metrics analysis#
df_subject_info.head(2)
| repositoryName | site | repository_and_site | subjectID | sessionID | run | age | gender | snr_total | slope | intercept | r_value | p_value | std_err | r_square | ICC | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| path | ||||||||||||||||
| IXI/Guys/sub-002/anat/sub-002_T1w.nii.gz | IXI | Guys | IXI|Guys | sub-002 | 1 | 1 | 3.58e+01 | FEMALE | 1.58e+01 | 9.76e-01 | 1.71e-01 | 8.71e-01 | 5.04e-47 | 4.54e-02 | 7.59e-01 | 8.66e-01 |
| IXI/Guys/sub-016/anat/sub-016_T1w.nii.gz | IXI | Guys | IXI|Guys | sub-016 | 1 | 1 | 5.52e+01 | MALE | 1.46e+01 | 1.07e+00 | -9.83e-02 | 9.00e-01 | 1.88e-54 | 4.28e-02 | 8.10e-01 | 8.87e-01 |
#@title
df_subject_info.groupby(by=var_compare).apply(lambda x: pd.Series([f'{x["ICC"].mean(): 0.2f}+/-{x["ICC"].std(): 0.2f}',
f'{x["r_square"].mean(): 0.2f}+/-{x["r_square"].std(): 0.2f}',
f'{x["slope"].mean(): 0.2f}+/-{x["slope"].std(): 0.2f}',
f'{x["intercept"].mean(): 0.2f}+/-{x["intercept"].std(): 0.2f}'],
index=['ICC', '$R^2$', 'slope', 'intercept']))
| ICC | $R^2$ | slope | intercept | |
|---|---|---|---|---|
| repository_and_site | ||||
| IXI|Guys | 0.82+/- 0.04 | 0.69+/- 0.06 | 0.95+/- 0.07 | 0.17+/- 0.17 |
| IXI|HH | 0.79+/- 0.07 | 0.65+/- 0.09 | 0.97+/- 0.12 | 0.09+/- 0.25 |
| IXI|IOP | 0.60+/- 0.08 | 0.38+/- 0.10 | 0.78+/- 0.12 | 0.54+/- 0.32 |
| OASIS3 | 0.80+/- 0.06 | 0.65+/- 0.08 | 0.89+/- 0.08 | 0.27+/- 0.19 |
def latex_float(float_str):
if "e" in float_str:
base, exponent = float_str.split("e")
return r"{0} \times 10^{{{1}}}".format(base, int(exponent))
else:
return float_str
#### Analysis of age effect on reproducibility metrics
import re, seaborn as sns
import numpy as np
from matplotlib import pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from matplotlib.colors import ListedColormap
from IPython.display import display, Math
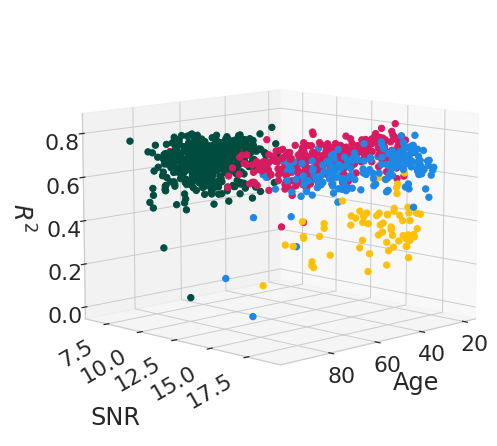
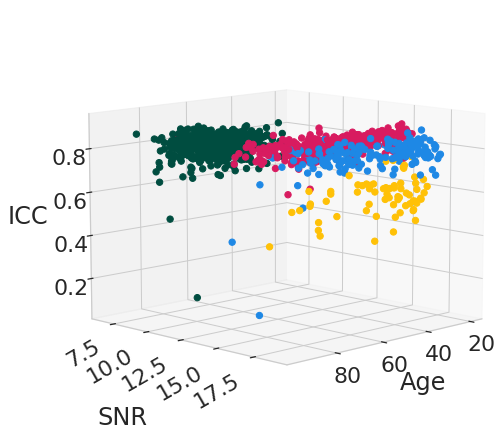
reproducibility_measures = [("r_square", "$R^2$"), ("ICC", "ICC")]
for rep_measure, label_measure in reproducibility_measures:
for el in df_subject_info[var_compare].unique():
legend = el
data_filtered = df_subject_info.loc[df_subject_info[var_compare] == el]
x_values = data_filtered[["age", "snr_total"]]
y_values = data_filtered[rep_measure]
model = LinearRegression().fit(x_values, y_values)
age_weight = latex_float(f'{model.coef_[0]: 0.1e}')
SNR_weight = latex_float(f'{model.coef_[1]: 0.1e}')
intercept = latex_float(f'{model.intercept_: 0.1e}')
eq = f'${label_measure.replace("$", "")}_' + "{" + legend + "}" + f'={age_weight}age + {SNR_weight}SNR + {intercept}$'
display(Math(r'{}'.format(eq)))
x= df_subject_info["age"].values
y= df_subject_info["snr_total"].values
z= df_subject_info[rep_measure].values
# axes instance
fig = plt.figure(figsize=(6,6))
ax = Axes3D(fig, auto_add_to_figure=False)
fig.add_axes(ax)
# get colormap from seaborn
cmap = ListedColormap(sns.color_palette("husl", 256).as_hex())
# plot
sc = ax.scatter(x, y, z, s=40, c=[color_pallete[el] for el in df_subject_info[var_compare].values], marker='o', cmap=cmap, alpha=1)
ax.set_xlabel('Age', labelpad=15)
ax.set_ylabel('SNR', labelpad=45)
ax.set_zlabel(label_measure, labelpad=15)
ax.view_init(elev=10., azim=45)
plt.subplots_adjust(bottom=0.2, top=2)
plt.setp(ax.get_yticklabels(), rotation=30, horizontalalignment='right')
plt.show()
\[\displaystyle R^2_{IXI|Guys}=-8.5 \times 10^{-4}age + 3.5 \times 10^{-3}SNR + 6.9 \times 10^{-1}\]
\[\displaystyle R^2_{IXI|HH}=-3.3 \times 10^{-4}age + 3.0 \times 10^{-2}SNR + 1.7 \times 10^{-1}\]
\[\displaystyle R^2_{IXI|IOP}=-3.8 \times 10^{-4}age + 2.2 \times 10^{-2}SNR + 2.4 \times 10^{-2}\]
\[\displaystyle R^2_{OASIS3}= 9.7 \times 10^{-4}age + 2.3 \times 10^{-2}SNR + 3.7 \times 10^{-1}\]
\[\displaystyle ICC_{IXI|Guys}=-3.2 \times 10^{-4}age + 2.6 \times 10^{-3}SNR + 8.0 \times 10^{-1}\]
\[\displaystyle ICC_{IXI|HH}= 1.1 \times 10^{-4}age + 2.6 \times 10^{-2}SNR + 3.6 \times 10^{-1}\]
\[\displaystyle ICC_{IXI|IOP}=-2.2 \times 10^{-4}age + 1.8 \times 10^{-2}SNR + 3.0 \times 10^{-1}\]
\[\displaystyle ICC_{OASIS3}= 9.1 \times 10^{-4}age + 1.9 \times 10^{-2}SNR + 5.6 \times 10^{-1}\]
ROI Analysis#

Compute metrics#
#@title
roi_grouped_by, roi_grouped_by_stats = get_regression_metrics(df_software, ["roiName", var_compare], (metric_analysis, pipeline1), (metric_analysis, pipeline2))
all_rois = []
for var in df_ct[var_compare].unique():
for roi_name in df_ct["roiName"].unique():
df_icc_roi = pg.intraclass_corr(data=df_ct.loc[(df_ct["roiName"] == roi_name) & (df_ct[var_compare] == var)],
targets='path', raters='software',
ratings='value')
df_icc_roi["roiName"] = roi_name
df_icc_roi[var_compare] = var
all_rois.append(df_icc_roi.loc[df_icc_roi["Type"]=="ICC3"])
df_icc_rois = pd.concat(all_rois)
roi_grouped_by_stats = roi_grouped_by_stats.join(pd.concat(all_rois).set_index(["roiName", var_compare])[["ICC", "CI95%"]])
map_abbrev_to_name = df_names_rois.set_index("label")[["name"]].to_dict("series")["name"]
hem_label = {"r": "Right", "l": "Left"}
roi_grouped_by_stats["name"] = roi_grouped_by_stats.index.to_frame().roiName.apply(lambda x: map_abbrev_to_name[x[1:]])
roi_grouped_by_stats["hem"] = roi_grouped_by_stats.index.to_frame().roiName.apply(lambda x: hem_label[x[0].lower()])
dfs = []
rois_info = roi_grouped_by_stats.copy()
rois_info["label"] = roi_grouped_by_stats.index.get_level_values(0).str[1:].str.strip()
rois_info["hem"] = roi_grouped_by_stats.index.get_level_values(0).str[0]
rois_info = rois_info.reset_index().set_index(["label", "hem", var_compare])
for var in rois_info.index.get_level_values(2).unique():
for hem in rois_info.index.get_level_values(1).unique():
df_areas_metrics_var = pd.concat([df_areas, df_areas["Label"].apply(lambda x: rois_info.loc[(x, hem, var)])], axis=1)
df_areas_metrics_var[var_compare] = var
dfs.append(df_areas_metrics_var)
df_areas_metrics = pd.concat(dfs)
df_areas_metrics["hem"] = df_areas_metrics["roiName"].str[0]
df_icc = df_areas_metrics.pivot_table(index="name", columns=[var_compare, "hem"], values=["ICC"])
df_ci = df_areas_metrics.pivot_table(index="name", columns=[var_compare, "hem"], values=["CI95%"])
df_ci_icc = df_icc.join(df_ci)
df_ci_icc.columns = df_ci_icc.columns.swaplevel(0, 2)
df_ci_icc.columns = df_ci_icc.columns.swaplevel(0, 1)
df_ci_icc = df_ci_icc[[df_ci_icc.columns[0], df_ci_icc.columns[-8],
df_ci_icc.columns[1], df_ci_icc.columns[-7],
df_ci_icc.columns[2], df_ci_icc.columns[-6],
df_ci_icc.columns[3], df_ci_icc.columns[-5],
df_ci_icc.columns[4], df_ci_icc.columns[-4],
df_ci_icc.columns[5], df_ci_icc.columns[-3],
df_ci_icc.columns[6], df_ci_icc.columns[-2],
df_ci_icc.columns[7], df_ci_icc.columns[-1]]]
roi_grouped_by_stats
| slope | intercept | r_value | p_value | std_err | r_square | ICC | CI95% | name | hem | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| roiName | repository_and_site | ||||||||||
| lG_Ins_lg_and_S_cent_ins | IXI|Guys | 4.43e-01 | 1.71e+00 | 4.91e-01 | 2.44e-20 | 4.46e-02 | 2.41e-01 | 4.88e-01 | [0.4, 0.57] | Long insular gyrus and central sulcus of the i... | Left |
| IXI|HH | 2.48e-01 | 2.13e+00 | 2.69e-01 | 2.76e-04 | 6.69e-02 | 7.22e-02 | 2.68e-01 | [0.13, 0.4] | Long insular gyrus and central sulcus of the i... | Left | |
| IXI|IOP | 2.29e-01 | 2.09e+00 | 2.97e-01 | 1.46e-02 | 9.12e-02 | 8.83e-02 | 2.87e-01 | [0.05, 0.49] | Long insular gyrus and central sulcus of the i... | Left | |
| OASIS3 | 4.76e-01 | 1.62e+00 | 5.30e-01 | 5.94e-37 | 3.44e-02 | 2.81e-01 | 5.27e-01 | [0.46, 0.59] | Long insular gyrus and central sulcus of the i... | Left | |
| lG_and_S_cingul-Ant | IXI|Guys | 5.44e-01 | 1.28e+00 | 5.91e-01 | 9.88e-31 | 4.22e-02 | 3.49e-01 | 5.89e-01 | [0.51, 0.66] | Anterior part of the cingulate gyrus and sulcus | Left |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| rS_temporal_sup | OASIS3 | 8.28e-01 | 3.38e-01 | 8.24e-01 | 6.46e-123 | 2.57e-02 | 6.80e-01 | 8.24e-01 | [0.79, 0.85] | Superior temporal sulcus | Right |
| rS_temporal_transverse | IXI|Guys | 7.02e-01 | 7.90e-01 | 4.96e-01 | 8.15e-21 | 6.97e-02 | 2.46e-01 | 4.68e-01 | [0.38, 0.55] | Transverse temporal sulcus | Right |
| IXI|HH | 7.15e-01 | 7.17e-01 | 4.46e-01 | 4.06e-10 | 1.08e-01 | 1.99e-01 | 4.00e-01 | [0.27, 0.52] | Transverse temporal sulcus | Right | |
| IXI|IOP | 5.92e-01 | 1.12e+00 | 3.64e-01 | 2.46e-03 | 1.88e-01 | 1.33e-01 | 3.25e-01 | [0.09, 0.52] | Transverse temporal sulcus | Right | |
| OASIS3 | 7.91e-01 | 6.32e-01 | 5.63e-01 | 1.89e-42 | 5.25e-02 | 3.17e-01 | 5.32e-01 | [0.47, 0.59] | Transverse temporal sulcus | Right |
592 rows × 10 columns
import pingouin as pg
all_df = []
for var in df_ct[var_compare].unique():
for roiname in df_ct['roiName'].unique():
df_posthoc = pg.pairwise_ttests(data=df_ct.loc[(df_ct['roiName'] == roiname) & (df_ct[var_compare] == var)],
dv='value', within='software', subject='path',
parametric=True, padjust='fdr_bh', effsize='cohen')
# Pretty printing of table
df_posthoc['roiName'] = roiname
df_posthoc[var_compare] = var
all_df.append(df_posthoc)
df_f_p_value = pd.concat(all_df)
df_f_p_value.head(4)
| Contrast | A | B | Paired | Parametric | T | dof | alternative | p-unc | BF10 | cohen | roiName | repository_and_site | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | software | ACPC_CAT12 | FREESURFER | True | True | -7.13e-01 | 3.11e+02 | two-sided | 4.76e-01 | 0.082 | -4.08e-02 | lG_Ins_lg_and_S_cent_ins | IXI|Guys |
| 0 | software | ACPC_CAT12 | FREESURFER | True | True | -1.83e+01 | 3.11e+02 | two-sided | 1.85e-51 | 9.934e+47 | -9.42e-01 | lG_and_S_cingul-Ant | IXI|Guys |
| 0 | software | ACPC_CAT12 | FREESURFER | True | True | 7.73e+00 | 3.11e+02 | two-sided | 1.45e-13 | 3.845e+10 | 3.97e-01 | lG_and_S_cingul-Mid-Ant | IXI|Guys |
| 0 | software | ACPC_CAT12 | FREESURFER | True | True | 5.18e-01 | 3.11e+02 | two-sided | 6.05e-01 | 0.073 | 2.17e-02 | lG_and_S_cingul-Mid-Post | IXI|Guys |
#@title
df_sorted = pd.pivot_table(roi_grouped_by_stats.reset_index(), index="name", values=["r_square"], columns=[var_compare, "hem"])
df_sorted
| r_square | ||||||||
|---|---|---|---|---|---|---|---|---|
| repository_and_site | IXI|Guys | IXI|HH | IXI|IOP | OASIS3 | ||||
| hem | Left | Right | Left | Right | Left | Right | Left | Right |
| name | ||||||||
| Angular gyrus | 6.29e-01 | 6.28e-01 | 6.89e-01 | 6.09e-01 | 4.25e-01 | 3.62e-01 | 5.78e-01 | 6.87e-01 |
| Anterior occipital sulcus and preoccipital notch | 4.92e-01 | 5.44e-01 | 3.70e-01 | 5.21e-01 | 4.99e-01 | 3.88e-01 | 3.45e-01 | 5.30e-01 |
| Anterior part of the cingulate gyrus and sulcus | 3.49e-01 | 3.31e-01 | 3.20e-01 | 4.29e-01 | 1.13e-01 | 1.38e-01 | 1.87e-01 | 1.98e-01 |
| Anterior segment of the circular sulcus of the insula | 1.10e-01 | 2.46e-01 | 8.89e-02 | 1.70e-01 | 5.75e-02 | 1.25e-02 | 2.34e-01 | 2.22e-01 |
| Anterior transverse collateral sulcus | 3.67e-03 | 1.26e-02 | 1.49e-02 | 1.97e-02 | 2.35e-03 | 2.21e-04 | 5.20e-05 | 5.61e-03 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| Temporal pole | 1.77e-01 | 3.16e-01 | 4.99e-01 | 3.75e-01 | 1.85e-01 | 9.46e-02 | 2.09e-01 | 2.70e-01 |
| Transverse frontopolar gyri and sulci | 3.25e-01 | 4.72e-01 | 4.36e-01 | 5.17e-01 | 9.51e-02 | 2.84e-01 | 1.87e-01 | 1.25e-01 |
| Transverse temporal sulcus | 5.10e-01 | 2.46e-01 | 4.22e-01 | 1.99e-01 | 4.97e-01 | 1.33e-01 | 3.74e-01 | 3.17e-01 |
| Triangular part of the inferior frontal gyrus | 6.30e-01 | 6.75e-01 | 5.94e-01 | 5.83e-01 | 5.60e-01 | 4.84e-01 | 6.28e-01 | 2.83e-01 |
| Vertical ramus of the anterior segment of the lateral sulcus | 2.76e-01 | 2.35e-01 | 2.55e-01 | 2.40e-01 | 2.01e-01 | 1.89e-01 | 3.46e-01 | 1.78e-01 |
74 rows × 8 columns
Reproducibility metrics analysis#
#@title
roi_grouped_by_stats.groupby(by=var_compare).apply(lambda x: pd.Series([f'{x["ICC"].mean(): 0.2f}+/-{x["ICC"].std(): 0.2f}',
f'{x["r_square"].mean(): 0.2f}+/-{x["r_square"].std(): 0.2f}',
f'{x["slope"].mean(): 0.2f}+/-{x["slope"].std(): 0.2f}',
f'{x["intercept"].mean(): 0.2f}+/-{x["intercept"].std(): 0.2f}'],
index=['ICC', '$R^2$', 'slope', 'intercept']))
| ICC | $R^2$ | slope | intercept | |
|---|---|---|---|---|
| repository_and_site | ||||
| IXI|Guys | 0.60+/- 0.18 | 0.40+/- 0.19 | 0.68+/- 0.22 | 0.82+/- 0.63 |
| IXI|HH | 0.59+/- 0.18 | 0.39+/- 0.19 | 0.72+/- 0.23 | 0.67+/- 0.61 |
| IXI|IOP | 0.44+/- 0.18 | 0.24+/- 0.16 | 0.56+/- 0.25 | 1.09+/- 0.76 |
| OASIS3 | 0.59+/- 0.20 | 0.39+/- 0.21 | 0.64+/- 0.23 | 0.88+/- 0.59 |
Mean per lobe#
#@title
to_group_by = ["Area"]
df_areas_metrics_grouped = df_areas_metrics.groupby(to_group_by + [var_compare, "hem"]).mean()
df_lobes = pd.pivot_table(df_areas_metrics_grouped.reset_index(), index=to_group_by, values=["r_square"], columns=[var_compare])
df_lobes
| r_square | ||||
|---|---|---|---|---|
| repository_and_site | IXI|Guys | IXI|HH | IXI|IOP | OASIS3 |
| Area | ||||
| Frontal Lobe | 4.01e-01 | 4.18e-01 | 2.55e-01 | 3.31e-01 |
| Insula | 3.42e-01 | 2.34e-01 | 2.02e-01 | 3.65e-01 |
| Limbic lobe | 3.23e-01 | 3.23e-01 | 1.90e-01 | 3.11e-01 |
| Parietal lobe | 5.14e-01 | 4.92e-01 | 2.99e-01 | 5.69e-01 |
| Temporal and occipital lobes | 4.15e-01 | 4.26e-01 | 2.49e-01 | 4.21e-01 |
Paired t-test#
all_df = []
for var in df_ct[var_compare].unique():
for roiname in df_ct['roiName'].unique():
df_posthoc = pg.pairwise_ttests(data=df_ct.loc[(df_ct['roiName'] == roiname) & (df_ct[var_compare] == var)],
dv='value', within='software', subject='path',
parametric=True, padjust='fdr_bh', effsize='cohen')
# Pretty printing of table
df_posthoc['roiName'] = roiname
df_posthoc[var_compare] = var
all_df.append(df_posthoc)
df_f_p_value = pd.concat(all_df)
df_f_p_value.head(4)
| Contrast | A | B | Paired | Parametric | T | dof | alternative | p-unc | BF10 | cohen | roiName | repository_and_site | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | software | ACPC_CAT12 | FREESURFER | True | True | -7.13e-01 | 3.11e+02 | two-sided | 4.76e-01 | 0.082 | -4.08e-02 | lG_Ins_lg_and_S_cent_ins | IXI|Guys |
| 0 | software | ACPC_CAT12 | FREESURFER | True | True | -1.83e+01 | 3.11e+02 | two-sided | 1.85e-51 | 9.934e+47 | -9.42e-01 | lG_and_S_cingul-Ant | IXI|Guys |
| 0 | software | ACPC_CAT12 | FREESURFER | True | True | 7.73e+00 | 3.11e+02 | two-sided | 1.45e-13 | 3.845e+10 | 3.97e-01 | lG_and_S_cingul-Mid-Ant | IXI|Guys |
| 0 | software | ACPC_CAT12 | FREESURFER | True | True | 5.18e-01 | 3.11e+02 | two-sided | 6.05e-01 | 0.073 | 2.17e-02 | lG_and_S_cingul-Mid-Post | IXI|Guys |
df_f_p_value["hem"] = df_f_p_value.apply(lambda x: hem_label[x.roiName[0].lower()], axis=1)
df_f_p_value["name"] = df_f_p_value.apply(lambda x: map_abbrev_to_name[x.roiName[1:]], axis=1)
# Bonferroni correction
significance_level = .05
df_f_p_value["p-value"] = df_f_p_value["p-unc"].apply(lambda x: min(x*df_f_p_value.shape[0], 1))
df_f_p_value_rejected = df_f_p_value[df_f_p_value["p-value"] < significance_level]
df_f_p_value["cohen"] = df_f_p_value["cohen"].astype(float).round(2).abs()
df_f_p_value["cohen-p-value"] = df_f_p_value[["p-value", "cohen"]].apply(lambda x: f'{x["cohen"]}*' if x['p-value'] < significance_level else f'{x["cohen"]}', axis=1).astype(str)
df_p_value = pd.pivot_table(df_f_p_value.reset_index(), index="name", values=["cohen-p-value"], columns=[var_compare, "hem"], aggfunc=lambda x: ' '.join(x))
df_p_value
| cohen-p-value | ||||||||
|---|---|---|---|---|---|---|---|---|
| repository_and_site | IXI|Guys | IXI|HH | IXI|IOP | OASIS3 | ||||
| hem | Left | Right | Left | Right | Left | Right | Left | Right |
| name | ||||||||
| Angular gyrus | 0.73* | 0.88* | 0.5* | 0.55* | 0.22 | 0.34 | 0.25* | 0.69* |
| Anterior occipital sulcus and preoccipital notch | 0.02 | 0.02 | 0.09 | 0.31* | 0.01 | 0.13 | 0.09 | 0.28* |
| Anterior part of the cingulate gyrus and sulcus | 0.94* | 0.73* | 0.81* | 0.48* | 0.64* | 0.56 | 0.21* | 0.44* |
| Anterior segment of the circular sulcus of the insula | 0.82* | 0.11 | 0.17 | 0.42* | 0.48 | 0.42 | 1.14* | 0.89* |
| Anterior transverse collateral sulcus | 0.67* | 0.48* | 0.88* | 0.85* | 2.38* | 1.85* | 0.19 | 0.44* |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| Temporal pole | 1.19* | 1.35* | 1.06* | 1.1* | 1.91* | 2.04* | 0.83* | 0.95* |
| Transverse frontopolar gyri and sulci | 0.76* | 0.35* | 0.12 | 0.0 | 0.45 | 0.25 | 0.65* | 0.51* |
| Transverse temporal sulcus | 0.25* | 0.57* | 0.04 | 0.33* | 0.17 | 0.26 | 0.45* | 0.75* |
| Triangular part of the inferior frontal gyrus | 0.82* | 0.82* | 0.54* | 0.62* | 0.42* | 0.65* | 0.28* | 0.11 |
| Vertical ramus of the anterior segment of the lateral sulcus | 1.09* | 0.42* | 1.07* | 0.31* | 1.53* | 1.4* | 1.32* | 1.12* |
74 rows × 8 columns
##### Number of regions per site in which the hypothesis was rejected
#@title
print(f"Number of ROIs with at least on group that rejected the paired t-test hypothesis: {df_f_p_value_rejected.reset_index().roiName.unique().shape[0]}")
Number of ROIs with at least on group that rejected the paired t-test hypothesis: 145
#@title
df_f_p_value_rejected.reset_index().groupby(var_compare).count()[["name"]]
| name | |
|---|---|
| repository_and_site | |
| IXI|Guys | 132 |
| IXI|HH | 116 |
| IXI|IOP | 94 |
| OASIS3 | 130 |
Modelling age using cortical thickness values#
df_cortical = pd.pivot_table(df_software_raw, index="path", columns=["software", "template", "roiName"],
values=df_software_raw.loc[:, ~df_software_raw.columns.isin(["software", "template", "roiName", "path", "age", "run"])])
df_age = df_subjects[["age", "gender", var_compare]]
df_age.columns=pd.MultiIndex.from_tuples([("age", "", "", ""), ("gender", "", "", ""), (var_compare, "", "", "")])
df_feat_raw = df_age.join(df_cortical).dropna().sample(frac=1)
import statsmodels.api as sm
from scipy import stats
map_df_p = {}
for soft in ["FREESURFER", "ACPC_CAT12"]:
X = df_feat_raw[("corticalThicknessAverage", soft, "a2009s")]
#X = X.rename(columns={el: f'{hem_names[el[0]]} {df_names_rois.loc[df_names_rois["label"] == el[1:]]["name"].iloc[0]}' for el in X.columns})
y = df_feat_raw["age"]
X2 = sm.add_constant(X)
est = sm.OLS(y, X2)
est2 = est.fit()
print(est2.summary())
map_df_p[soft] = pd.DataFrame(est2.summary().tables[1].data[1:], columns=est2.summary().tables[1].data[0]).rename(columns={"": "roiName"})
OLS Regression Results
==============================================================================
Dep. Variable: age R-squared: 0.750
Model: OLS Adj. R-squared: 0.709
Method: Least Squares F-statistic: 18.25
Date: Wed, 31 Aug 2022 Prob (F-statistic): 3.28e-190
Time: 18:57:25 Log-Likelihood: -3706.6
No. Observations: 1048 AIC: 7711.
Df Residuals: 899 BIC: 8449.
Df Model: 148
Covariance Type: nonrobust
==============================================================================================
coef std err t P>|t| [0.025 0.975]
----------------------------------------------------------------------------------------------
const 162.6633 9.607 16.932 0.000 143.809 181.517
lG_Ins_lg_and_S_cent_ins 1.8801 1.641 1.146 0.252 -1.340 5.100
lG_and_S_cingul-Ant 4.5904 3.015 1.523 0.128 -1.327 10.507
lG_and_S_cingul-Mid-Ant -1.7949 2.650 -0.677 0.498 -6.995 3.405
lG_and_S_cingul-Mid-Post 5.8002 3.221 1.801 0.072 -0.521 12.121
lG_and_S_frontomargin 2.8552 2.409 1.185 0.236 -1.873 7.583
lG_and_S_occipital_inf 2.7777 2.210 1.257 0.209 -1.559 7.114
lG_and_S_paracentral 2.6367 2.489 1.059 0.290 -2.248 7.522
lG_and_S_subcentral -2.3093 2.655 -0.870 0.385 -7.520 2.902
lG_and_S_transv_frontopol -2.8722 2.117 -1.357 0.175 -7.027 1.282
lG_cingul-Post-dorsal -3.1601 2.608 -1.212 0.226 -8.279 1.958
lG_cingul-Post-ventral -0.7145 1.508 -0.474 0.636 -3.675 2.246
lG_cuneus 5.5720 3.505 1.590 0.112 -1.306 12.450
lG_front_inf-Opercular 1.0811 2.909 0.372 0.710 -4.628 6.790
lG_front_inf-Orbital -2.1892 1.857 -1.179 0.239 -5.834 1.456
lG_front_inf-Triangul -6.1017 2.592 -2.354 0.019 -11.189 -1.014
lG_front_middle 0.0196 4.144 0.005 0.996 -8.114 8.153
lG_front_sup -13.6879 4.716 -2.903 0.004 -22.943 -4.433
lG_insular_short 2.0511 1.581 1.298 0.195 -1.051 5.153
lG_oc-temp_lat-fusifor 2.4719 2.424 1.020 0.308 -2.286 7.229
lG_oc-temp_med-Lingual 10.1881 3.884 2.623 0.009 2.565 17.811
lG_oc-temp_med-Parahip 1.4166 1.825 0.776 0.438 -2.166 4.999
lG_occipital_middle -2.9110 3.264 -0.892 0.373 -9.317 3.495
lG_occipital_sup -1.5066 2.536 -0.594 0.553 -6.484 3.471
lG_orbital -4.8121 3.059 -1.573 0.116 -10.816 1.192
lG_pariet_inf-Angular -0.3133 3.173 -0.099 0.921 -6.540 5.914
lG_pariet_inf-Supramar 4.6408 3.338 1.390 0.165 -1.911 11.193
lG_parietal_sup -1.6273 3.845 -0.423 0.672 -9.173 5.918
lG_postcentral 6.1216 3.115 1.965 0.050 0.008 12.235
lG_precentral -2.4811 2.685 -0.924 0.356 -7.750 2.788
lG_precuneus 3.5276 3.454 1.021 0.307 -3.252 10.307
lG_rectus -0.1284 2.189 -0.059 0.953 -4.425 4.168
lG_subcallosal -7.0937 1.605 -4.419 0.000 -10.244 -3.943
lG_temp_sup-G_T_transv 2.3114 1.772 1.305 0.192 -1.166 5.788
lG_temp_sup-Lateral 1.5336 2.593 0.592 0.554 -3.555 6.622
lG_temp_sup-Plan_polar -1.8756 1.576 -1.190 0.234 -4.968 1.217
lG_temp_sup-Plan_tempo -0.6424 2.186 -0.294 0.769 -4.932 3.647
lG_temporal_inf -0.4700 2.750 -0.171 0.864 -5.867 4.927
lG_temporal_middle 0.1601 2.927 0.055 0.956 -5.585 5.905
lLat_Fis-ant-Horizont 2.0188 1.392 1.451 0.147 -0.713 4.750
lLat_Fis-ant-Vertical -1.3088 1.477 -0.886 0.376 -4.208 1.590
lLat_Fis-post -4.6103 2.637 -1.748 0.081 -9.786 0.565
lPole_occipital 4.5192 2.968 1.523 0.128 -1.305 10.343
lPole_temporal 3.4046 1.988 1.713 0.087 -0.497 7.306
lS_calcarine -6.7020 3.972 -1.687 0.092 -14.497 1.093
lS_central 2.2414 3.620 0.619 0.536 -4.864 9.347
lS_cingul-Marginalis -3.8176 2.916 -1.309 0.191 -9.541 1.906
lS_circular_insula_ant -1.5289 1.790 -0.854 0.393 -5.041 1.984
lS_circular_insula_inf -1.1256 2.191 -0.514 0.608 -5.426 3.175
lS_circular_insula_sup -7.2360 3.542 -2.043 0.041 -14.188 -0.284
lS_collat_transv_ant 1.9131 1.589 1.204 0.229 -1.205 5.031
lS_collat_transv_post -3.3025 2.076 -1.591 0.112 -7.376 0.771
lS_front_inf -0.1988 3.695 -0.054 0.957 -7.450 7.052
lS_front_middle -2.4258 3.015 -0.805 0.421 -8.343 3.491
lS_front_sup 0.7728 3.837 0.201 0.840 -6.758 8.304
lS_interm_prim-Jensen 0.0441 1.090 0.040 0.968 -2.096 2.184
lS_intrapariet_and_P_trans 5.5417 3.293 1.683 0.093 -0.921 12.004
lS_oc-temp_lat -2.8196 2.153 -1.310 0.191 -7.044 1.405
lS_oc-temp_med_and_Lingual -5.8314 2.859 -2.039 0.042 -11.443 -0.220
lS_oc_middle_and_Lunatus 1.3533 2.410 0.562 0.574 -3.376 6.082
lS_oc_sup_and_transversal 1.2080 3.207 0.377 0.706 -5.086 7.501
lS_occipital_ant 1.8708 2.214 0.845 0.398 -2.475 6.217
lS_orbital-H_Shaped 4.9365 2.056 2.401 0.017 0.902 8.971
lS_orbital_lateral -0.7144 1.585 -0.451 0.652 -3.825 2.396
lS_orbital_med-olfact -4.2319 1.787 -2.368 0.018 -7.739 -0.725
lS_parieto_occipital -0.8894 2.988 -0.298 0.766 -6.753 4.975
lS_pericallosal 1.3095 1.791 0.731 0.465 -2.205 4.824
lS_postcentral -4.8329 3.797 -1.273 0.203 -12.285 2.620
lS_precentral-inf-part 1.3758 3.033 0.454 0.650 -4.576 7.328
lS_precentral-sup-part -3.4558 2.399 -1.441 0.150 -8.164 1.252
lS_suborbital -2.3545 1.564 -1.505 0.133 -5.425 0.716
lS_subparietal -0.8569 2.720 -0.315 0.753 -6.196 4.482
lS_temporal_inf -1.0085 2.565 -0.393 0.694 -6.042 4.025
lS_temporal_sup -6.4322 4.668 -1.378 0.169 -15.593 2.729
lS_temporal_transverse -0.7816 1.255 -0.623 0.534 -3.244 1.681
rG_Ins_lg_and_S_cent_ins 2.9556 1.346 2.195 0.028 0.313 5.598
rG_and_S_cingul-Ant 4.1536 3.124 1.330 0.184 -1.978 10.285
rG_and_S_cingul-Mid-Ant -5.3265 2.928 -1.819 0.069 -11.072 0.419
rG_and_S_cingul-Mid-Post 2.6477 3.295 0.804 0.422 -3.819 9.114
rG_and_S_frontomargin 3.7501 2.035 1.843 0.066 -0.243 7.743
rG_and_S_occipital_inf 4.4235 1.828 2.420 0.016 0.837 8.010
rG_and_S_paracentral -5.7832 2.389 -2.420 0.016 -10.473 -1.094
rG_and_S_subcentral -2.2775 2.486 -0.916 0.360 -7.156 2.601
rG_and_S_transv_frontopol 0.7548 2.394 0.315 0.753 -3.944 5.454
rG_cingul-Post-dorsal -2.9330 2.376 -1.234 0.217 -7.597 1.731
rG_cingul-Post-ventral 0.2480 1.683 0.147 0.883 -3.055 3.551
rG_cuneus 3.2595 3.669 0.888 0.375 -3.942 10.461
rG_front_inf-Opercular -0.9917 2.838 -0.349 0.727 -6.561 4.578
rG_front_inf-Orbital 1.1768 1.769 0.665 0.506 -2.295 4.648
rG_front_inf-Triangul -1.1011 2.409 -0.457 0.648 -5.829 3.626
rG_front_middle 6.2971 3.817 1.650 0.099 -1.194 13.788
rG_front_sup -0.7769 4.769 -0.163 0.871 -10.136 8.582
rG_insular_short 1.3954 1.394 1.001 0.317 -1.341 4.132
rG_oc-temp_lat-fusifor 2.3353 2.456 0.951 0.342 -2.486 7.156
rG_oc-temp_med-Lingual 9.7579 3.494 2.793 0.005 2.901 16.615
rG_oc-temp_med-Parahip 3.5787 1.883 1.901 0.058 -0.116 7.274
rG_occipital_middle -1.2176 3.435 -0.354 0.723 -7.959 5.524
rG_occipital_sup -5.0534 2.799 -1.806 0.071 -10.546 0.440
rG_orbital -3.0277 2.906 -1.042 0.298 -8.731 2.676
rG_pariet_inf-Angular -9.8900 3.534 -2.799 0.005 -16.825 -2.955
rG_pariet_inf-Supramar 4.4915 3.400 1.321 0.187 -2.182 11.165
rG_parietal_sup -1.1754 3.331 -0.353 0.724 -7.713 5.362
rG_postcentral -4.2514 2.873 -1.480 0.139 -9.890 1.387
rG_precentral -0.3874 2.537 -0.153 0.879 -5.366 4.591
rG_precuneus 4.7898 3.189 1.502 0.133 -1.470 11.049
rG_rectus -2.9699 1.973 -1.505 0.133 -6.842 0.902
rG_subcallosal 1.6878 1.608 1.050 0.294 -1.467 4.843
rG_temp_sup-G_T_transv 3.4824 1.760 1.979 0.048 0.028 6.936
rG_temp_sup-Lateral -5.5556 2.598 -2.139 0.033 -10.654 -0.457
rG_temp_sup-Plan_polar -2.2680 1.539 -1.474 0.141 -5.288 0.752
rG_temp_sup-Plan_tempo 0.6442 2.243 0.287 0.774 -3.758 5.047
rG_temporal_inf 2.8384 2.507 1.132 0.258 -2.081 7.758
rG_temporal_middle -4.6515 3.245 -1.433 0.152 -11.020 1.717
rLat_Fis-ant-Horizont -2.8900 1.604 -1.801 0.072 -6.039 0.259
rLat_Fis-ant-Vertical -2.8662 1.319 -2.173 0.030 -5.455 -0.278
rLat_Fis-post -3.8852 2.928 -1.327 0.185 -9.632 1.862
rPole_occipital 10.1364 3.558 2.849 0.004 3.154 17.119
rPole_temporal 4.2463 2.067 2.054 0.040 0.189 8.304
rS_calcarine -6.8774 3.641 -1.889 0.059 -14.024 0.269
rS_central 15.9589 3.721 4.289 0.000 8.656 23.262
rS_cingul-Marginalis 0.0957 3.132 0.031 0.976 -6.052 6.243
rS_circular_insula_ant -4.8856 1.695 -2.883 0.004 -8.212 -1.559
rS_circular_insula_inf -10.7969 2.090 -5.167 0.000 -14.898 -6.696
rS_circular_insula_sup -6.5451 3.239 -2.020 0.044 -12.903 -0.188
rS_collat_transv_ant -0.8794 1.613 -0.545 0.586 -4.046 2.287
rS_collat_transv_post 0.0118 2.062 0.006 0.995 -4.034 4.058
rS_front_inf 2.1303 3.471 0.614 0.540 -4.682 8.943
rS_front_middle 0.7142 3.278 0.218 0.828 -5.718 7.147
rS_front_sup -6.7213 3.816 -1.761 0.079 -14.211 0.769
rS_interm_prim-Jensen -1.3392 1.604 -0.835 0.404 -4.487 1.808
rS_intrapariet_and_P_trans 4.6399 4.084 1.136 0.256 -3.376 12.656
rS_oc-temp_lat 1.2472 1.937 0.644 0.520 -2.554 5.049
rS_oc-temp_med_and_Lingual -7.0609 2.794 -2.527 0.012 -12.544 -1.578
rS_oc_middle_and_Lunatus -3.9183 2.388 -1.641 0.101 -8.605 0.768
rS_oc_sup_and_transversal 7.9981 2.938 2.722 0.007 2.232 13.764
rS_occipital_ant -4.8187 2.128 -2.264 0.024 -8.995 -0.642
rS_orbital-H_Shaped 3.2383 2.130 1.520 0.129 -0.942 7.419
rS_orbital_lateral -5.6681 1.693 -3.348 0.001 -8.991 -2.345
rS_orbital_med-olfact -1.0335 1.755 -0.589 0.556 -4.478 2.411
rS_parieto_occipital 2.1148 3.251 0.651 0.516 -4.266 8.495
rS_pericallosal 3.6538 1.657 2.205 0.028 0.402 6.906
rS_postcentral 4.4598 3.438 1.297 0.195 -2.288 11.208
rS_precentral-inf-part 0.3042 2.953 0.103 0.918 -5.492 6.101
rS_precentral-sup-part -5.6903 2.430 -2.341 0.019 -10.460 -0.920
rS_suborbital -1.7873 0.974 -1.835 0.067 -3.698 0.124
rS_subparietal 2.2839 2.675 0.854 0.393 -2.965 7.533
rS_temporal_inf -0.2761 2.528 -0.109 0.913 -5.238 4.686
rS_temporal_sup 6.7630 4.679 1.445 0.149 -2.420 15.946
rS_temporal_transverse -2.8381 1.178 -2.410 0.016 -5.150 -0.527
==============================================================================
Omnibus: 1.996 Durbin-Watson: 2.026
Prob(Omnibus): 0.369 Jarque-Bera (JB): 2.069
Skew: -0.096 Prob(JB): 0.355
Kurtosis: 2.897 Cond. No. 1.04e+03
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[2] The condition number is large, 1.04e+03. This might indicate that there are
strong multicollinearity or other numerical problems.
OLS Regression Results
==============================================================================
Dep. Variable: age R-squared: 0.769
Model: OLS Adj. R-squared: 0.731
Method: Least Squares F-statistic: 20.24
Date: Wed, 31 Aug 2022 Prob (F-statistic): 9.85e-205
Time: 18:57:25 Log-Likelihood: -3665.5
No. Observations: 1048 AIC: 7629.
Df Residuals: 899 BIC: 8367.
Df Model: 148
Covariance Type: nonrobust
==============================================================================================
coef std err t P>|t| [0.025 0.975]
----------------------------------------------------------------------------------------------
const 130.9484 11.135 11.760 0.000 109.094 152.803
lG_Ins_lg_and_S_cent_ins 2.6798 1.377 1.946 0.052 -0.023 5.383
lG_and_S_cingul-Ant -1.1407 2.441 -0.467 0.640 -5.931 3.650
lG_and_S_cingul-Mid-Ant 1.1020 2.447 0.450 0.653 -3.701 5.905
lG_and_S_cingul-Mid-Post -1.3026 2.999 -0.434 0.664 -7.188 4.583
lG_and_S_frontomargin -4.3803 2.881 -1.520 0.129 -10.035 1.274
lG_and_S_occipital_inf 3.0928 2.296 1.347 0.178 -1.414 7.599
lG_and_S_paracentral 9.8718 3.250 3.037 0.002 3.492 16.251
lG_and_S_subcentral 6.9062 3.591 1.923 0.055 -0.141 13.953
lG_and_S_transv_frontopol 2.4260 2.452 0.989 0.323 -2.386 7.238
lG_cingul-Post-dorsal 2.7937 1.968 1.419 0.156 -1.069 6.656
lG_cingul-Post-ventral 0.5031 1.338 0.376 0.707 -2.123 3.130
lG_cuneus -10.6294 5.027 -2.115 0.035 -20.495 -0.764
lG_front_inf-Opercular -6.8663 3.558 -1.930 0.054 -13.848 0.116
lG_front_inf-Orbital 1.1076 1.543 0.718 0.473 -1.920 4.136
lG_front_inf-Triangul -8.6061 3.366 -2.557 0.011 -15.212 -2.000
lG_front_middle -7.0279 5.205 -1.350 0.177 -17.242 3.187
lG_front_sup -13.8983 5.282 -2.631 0.009 -24.265 -3.531
lG_insular_short 1.5305 1.039 1.473 0.141 -0.508 3.569
lG_oc-temp_lat-fusifor 4.8297 2.577 1.874 0.061 -0.228 9.887
lG_oc-temp_med-Lingual 9.9359 4.348 2.285 0.023 1.402 18.470
lG_oc-temp_med-Parahip -1.1773 1.785 -0.660 0.510 -4.681 2.326
lG_occipital_middle -4.3728 3.708 -1.179 0.239 -11.649 2.904
lG_occipital_sup -11.2158 3.737 -3.001 0.003 -18.551 -3.881
lG_orbital 4.0717 3.029 1.344 0.179 -1.872 10.016
lG_pariet_inf-Angular -1.8394 4.162 -0.442 0.659 -10.008 6.330
lG_pariet_inf-Supramar 6.2437 4.741 1.317 0.188 -3.062 15.549
lG_parietal_sup -9.5152 4.616 -2.061 0.040 -18.574 -0.456
lG_postcentral 0.0385 3.795 0.010 0.992 -7.410 7.487
lG_precentral 6.8333 3.709 1.842 0.066 -0.447 14.113
lG_precuneus 12.8999 4.295 3.003 0.003 4.471 21.329
lG_rectus -6.6429 2.695 -2.465 0.014 -11.932 -1.354
lG_subcallosal 0.0105 1.032 0.010 0.992 -2.015 2.036
lG_temp_sup-G_T_transv -13.5879 2.435 -5.580 0.000 -18.367 -8.809
lG_temp_sup-Lateral 15.5119 2.457 6.312 0.000 10.689 20.335
lG_temp_sup-Plan_polar -1.7194 1.522 -1.129 0.259 -4.707 1.268
lG_temp_sup-Plan_tempo -7.1905 2.966 -2.425 0.016 -13.011 -1.370
lG_temporal_inf 5.0976 2.805 1.817 0.070 -0.408 10.603
lG_temporal_middle 3.0762 2.996 1.027 0.305 -2.803 8.955
lLat_Fis-ant-Horizont 0.0620 1.332 0.047 0.963 -2.552 2.676
lLat_Fis-ant-Vertical 1.2540 2.113 0.594 0.553 -2.892 5.400
lLat_Fis-post -4.7467 3.142 -1.511 0.131 -10.913 1.420
lPole_occipital 9.4395 4.227 2.233 0.026 1.144 17.735
lPole_temporal -0.1191 1.820 -0.065 0.948 -3.690 3.452
lS_calcarine -7.4732 3.487 -2.143 0.032 -14.317 -0.630
lS_central -28.0741 5.301 -5.296 0.000 -38.478 -17.670
lS_cingul-Marginalis -5.2068 3.110 -1.674 0.094 -11.311 0.897
lS_circular_insula_ant -1.9856 1.549 -1.282 0.200 -5.026 1.055
lS_circular_insula_inf -1.9640 2.205 -0.891 0.373 -6.291 2.363
lS_circular_insula_sup -2.2063 3.656 -0.603 0.546 -9.382 4.969
lS_collat_transv_ant 2.5332 1.575 1.608 0.108 -0.558 5.624
lS_collat_transv_post -1.2532 2.262 -0.554 0.580 -5.692 3.185
lS_front_inf -0.8407 4.029 -0.209 0.835 -8.749 7.067
lS_front_middle 0.2424 3.163 0.077 0.939 -5.966 6.451
lS_front_sup 1.2222 4.170 0.293 0.770 -6.962 9.406
lS_interm_prim-Jensen -0.4008 1.683 -0.238 0.812 -3.703 2.902
lS_intrapariet_and_P_trans 4.0641 4.463 0.911 0.363 -4.696 12.824
lS_oc-temp_lat -0.7334 1.967 -0.373 0.709 -4.594 3.128
lS_oc-temp_med_and_Lingual -2.3131 2.578 -0.897 0.370 -7.373 2.747
lS_oc_middle_and_Lunatus 4.6986 2.783 1.688 0.092 -0.763 10.161
lS_oc_sup_and_transversal 6.9038 3.255 2.121 0.034 0.516 13.291
lS_occipital_ant 6.7092 2.143 3.131 0.002 2.504 10.915
lS_orbital-H_Shaped 4.3004 2.641 1.629 0.104 -0.882 9.483
lS_orbital_lateral 2.3065 2.071 1.114 0.266 -1.757 6.370
lS_orbital_med-olfact -3.7949 1.578 -2.405 0.016 -6.892 -0.697
lS_parieto_occipital -0.5678 3.519 -0.161 0.872 -7.475 6.339
lS_pericallosal 1.2107 2.267 0.534 0.593 -3.238 5.659
lS_postcentral 5.2739 3.988 1.323 0.186 -2.552 13.100
lS_precentral-inf-part 9.6395 3.478 2.772 0.006 2.814 16.465
lS_precentral-sup-part 0.4949 3.101 0.160 0.873 -5.591 6.580
lS_suborbital -0.7684 1.656 -0.464 0.643 -4.018 2.481
lS_subparietal 1.7525 3.060 0.573 0.567 -4.253 7.758
lS_temporal_inf -7.0254 2.436 -2.884 0.004 -11.807 -2.244
lS_temporal_sup -11.2065 4.091 -2.739 0.006 -19.236 -3.177
lS_temporal_transverse 3.8177 1.759 2.170 0.030 0.365 7.270
rG_Ins_lg_and_S_cent_ins 1.1106 1.183 0.939 0.348 -1.211 3.433
rG_and_S_cingul-Ant -2.6747 2.617 -1.022 0.307 -7.811 2.461
rG_and_S_cingul-Mid-Ant 0.8564 2.374 0.361 0.718 -3.803 5.516
rG_and_S_cingul-Mid-Post -2.6059 3.123 -0.834 0.404 -8.735 3.523
rG_and_S_frontomargin 0.5559 2.649 0.210 0.834 -4.643 5.755
rG_and_S_occipital_inf -0.2058 2.134 -0.096 0.923 -4.394 3.983
rG_and_S_paracentral 1.1009 2.930 0.376 0.707 -4.650 6.851
rG_and_S_subcentral -0.0187 3.228 -0.006 0.995 -6.354 6.316
rG_and_S_transv_frontopol 0.5471 2.725 0.201 0.841 -4.800 5.895
rG_cingul-Post-dorsal 0.9095 2.090 0.435 0.663 -3.192 5.011
rG_cingul-Post-ventral 1.8786 1.440 1.304 0.193 -0.948 4.706
rG_cuneus 20.2956 4.706 4.313 0.000 11.060 29.531
rG_front_inf-Opercular -0.4540 3.290 -0.138 0.890 -6.912 6.004
rG_front_inf-Orbital -0.7970 1.509 -0.528 0.598 -3.758 2.164
rG_front_inf-Triangul -0.3777 3.186 -0.119 0.906 -6.631 5.875
rG_front_middle -4.7727 4.804 -0.994 0.321 -14.200 4.655
rG_front_sup -17.1571 4.961 -3.458 0.001 -26.894 -7.420
rG_insular_short 0.3925 1.014 0.387 0.699 -1.598 2.384
rG_oc-temp_lat-fusifor -3.4502 2.520 -1.369 0.171 -8.397 1.496
rG_oc-temp_med-Lingual 3.1739 3.774 0.841 0.401 -4.233 10.581
rG_oc-temp_med-Parahip 2.1334 1.747 1.221 0.222 -1.296 5.562
rG_occipital_middle 9.7241 3.922 2.480 0.013 2.028 17.421
rG_occipital_sup -9.4860 3.732 -2.542 0.011 -16.811 -2.161
rG_orbital -5.9737 3.071 -1.945 0.052 -12.001 0.054
rG_pariet_inf-Angular -13.4195 4.020 -3.338 0.001 -21.310 -5.529
rG_pariet_inf-Supramar 4.1626 5.112 0.814 0.416 -5.870 14.195
rG_parietal_sup -2.1805 4.345 -0.502 0.616 -10.708 6.347
rG_postcentral -6.9020 3.570 -1.933 0.054 -13.908 0.104
rG_precentral -2.4905 3.558 -0.700 0.484 -9.474 4.493
rG_precuneus 8.0284 4.060 1.978 0.048 0.061 15.996
rG_rectus 3.4696 2.483 1.398 0.163 -1.403 8.342
rG_subcallosal 2.5969 1.049 2.475 0.013 0.538 4.656
rG_temp_sup-G_T_transv -12.4157 2.089 -5.942 0.000 -16.516 -8.315
rG_temp_sup-Lateral 3.4355 2.655 1.294 0.196 -1.775 8.646
rG_temp_sup-Plan_polar 2.1193 1.613 1.313 0.189 -1.047 5.286
rG_temp_sup-Plan_tempo -9.1953 2.874 -3.200 0.001 -14.835 -3.556
rG_temporal_inf 0.7054 2.620 0.269 0.788 -4.436 5.847
rG_temporal_middle -0.3558 3.404 -0.105 0.917 -7.037 6.325
rLat_Fis-ant-Horizont -3.1008 1.687 -1.838 0.066 -6.412 0.210
rLat_Fis-ant-Vertical -1.2958 1.563 -0.829 0.407 -4.364 1.772
rLat_Fis-post 9.4423 2.859 3.303 0.001 3.831 15.054
rPole_occipital 11.2538 4.340 2.593 0.010 2.736 19.772
rPole_temporal -2.1337 1.628 -1.310 0.190 -5.330 1.062
rS_calcarine -8.1572 3.350 -2.435 0.015 -14.733 -1.582
rS_central -6.5683 5.282 -1.244 0.214 -16.935 3.798
rS_cingul-Marginalis 4.1916 3.707 1.131 0.258 -3.083 11.466
rS_circular_insula_ant -0.5190 1.837 -0.282 0.778 -4.125 3.087
rS_circular_insula_inf -1.8573 1.957 -0.949 0.343 -5.697 1.983
rS_circular_insula_sup -5.0688 3.248 -1.560 0.119 -11.444 1.306
rS_collat_transv_ant -1.4828 1.596 -0.929 0.353 -4.614 1.649
rS_collat_transv_post 0.1848 2.355 0.078 0.937 -4.436 4.806
rS_front_inf -4.6702 4.045 -1.155 0.249 -12.609 3.269
rS_front_middle 6.4926 3.885 1.671 0.095 -1.132 14.117
rS_front_sup -5.2126 4.366 -1.194 0.233 -13.782 3.357
rS_interm_prim-Jensen 4.6899 2.517 1.863 0.063 -0.250 9.630
rS_intrapariet_and_P_trans -4.0581 4.494 -0.903 0.367 -12.878 4.762
rS_oc-temp_lat -0.2339 1.761 -0.133 0.894 -3.690 3.223
rS_oc-temp_med_and_Lingual -1.7311 2.537 -0.682 0.495 -6.711 3.249
rS_oc_middle_and_Lunatus -1.3691 2.482 -0.551 0.581 -6.241 3.503
rS_oc_sup_and_transversal 6.9599 3.199 2.176 0.030 0.682 13.238
rS_occipital_ant -0.7138 2.227 -0.320 0.749 -5.085 3.658
rS_orbital-H_Shaped 4.4708 2.829 1.580 0.114 -1.082 10.024
rS_orbital_lateral 0.3258 1.970 0.165 0.869 -3.540 4.191
rS_orbital_med-olfact 4.2423 1.826 2.323 0.020 0.658 7.827
rS_parieto_occipital 1.0090 3.560 0.283 0.777 -5.977 7.995
rS_pericallosal -0.8188 2.622 -0.312 0.755 -5.964 4.326
rS_postcentral 5.9814 3.678 1.626 0.104 -1.238 13.200
rS_precentral-inf-part 8.0933 3.627 2.232 0.026 0.976 15.211
rS_precentral-sup-part 10.5917 3.095 3.422 0.001 4.517 16.666
rS_suborbital -2.4750 1.381 -1.793 0.073 -5.185 0.235
rS_subparietal 2.8485 3.015 0.945 0.345 -3.069 8.766
rS_temporal_inf -2.4824 2.442 -1.017 0.310 -7.275 2.310
rS_temporal_sup -5.4213 4.509 -1.202 0.230 -14.271 3.428
rS_temporal_transverse 0.8230 1.896 0.434 0.664 -2.898 4.544
==============================================================================
Omnibus: 8.905 Durbin-Watson: 2.021
Prob(Omnibus): 0.012 Jarque-Bera (JB): 8.960
Skew: -0.211 Prob(JB): 0.0113
Kurtosis: 2.833 Cond. No. 1.23e+03
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[2] The condition number is large, 1.23e+03. This might indicate that there are
strong multicollinearity or other numerical problems.
/tmp/ipykernel_42420/2286560134.py:7: PerformanceWarning: indexing past lexsort depth may impact performance.
X = df_feat_raw[("corticalThicknessAverage", soft, "a2009s")]
/tmp/ipykernel_42420/2286560134.py:7: PerformanceWarning: indexing past lexsort depth may impact performance.
X = df_feat_raw[("corticalThicknessAverage", soft, "a2009s")]
areas_relation_age_fs = map_df_p["FREESURFER"].loc[map_df_p["FREESURFER"]["P>|t|"].astype(float) < 0.05]["roiName"].to_list()
areas_relation_age_cat12 = map_df_p["ACPC_CAT12"].loc[map_df_p["ACPC_CAT12"]["P>|t|"].astype(float) < 0.05]["roiName"].to_list()
areas_relation_age_fs.remove('const')
areas_relation_age_cat12.remove('const')
intersection_areas = list(set(areas_relation_age_fs) & set(areas_relation_age_cat12))
print("Overlap ROIs")
df_overlap = df_areas_metrics.loc[df_areas_metrics["roiName"].isin(intersection_areas)].groupby(["roiName", "name"]).mean()[["r_square"]]
df_overlap
Overlap ROIs
| r_square | ||
|---|---|---|
| roiName | name | |
| lG_front_inf-Triangul | Triangular part of the inferior frontal gyrus | 6.03e-01 |
| lG_front_sup | Superior frontal gyrus | 6.60e-01 |
| lG_oc-temp_med-Lingual | Lingual gyrus | 2.66e-01 |
| lS_orbital_med-olfact | Medial orbital sulcus | 2.67e-02 |
| rG_pariet_inf-Angular | Angular gyrus | 5.71e-01 |
| rG_temp_sup-G_T_transv | Anterior transverse temporal gyrus | 2.81e-01 |
| rPole_occipital | Occipital pole | 3.43e-01 |
| rS_oc_sup_and_transversal | Superior occipital sulcus and transverse occipital sulcus | 4.81e-01 |
| rS_precentral-sup-part | Superior part of the precentral sulcus | 4.31e-01 |
cat12_rois_important_non_overlap = [el for el in areas_relation_age_cat12 if el not in intersection_areas]
df_cat12_important = df_areas_metrics.loc[df_areas_metrics["roiName"].isin(cat12_rois_important_non_overlap)].groupby(["roiName", "name"]).mean()[["r_square"]]
fs_rois_important_non_overlap = [el for el in areas_relation_age_fs if el not in intersection_areas]
df_fs_important = df_areas_metrics.loc[df_areas_metrics["roiName"].isin(fs_rois_important_non_overlap)].groupby(["roiName", "name"]).mean()[["r_square"]]
display(HTML(f"<br><h5>FreeSurfer</h5>"))
display(HTML(df_fs_important.to_html()))
display(HTML(f"<br><h5>CAT12</h5>"))
display(HTML(df_cat12_important.to_html()))
FreeSurfer
| r_square | ||
|---|---|---|
| roiName | name | |
| lG_subcallosal | Subcallosal area | 2.40e-03 |
| lS_circular_insula_sup | Superior segment of the circular sulcus of the insula | 4.40e-01 |
| lS_oc-temp_med_and_Lingual | Medial occipito-temporal sulcus and lingual sulcus | 4.03e-01 |
| lS_orbital-H_Shaped | Orbital sulci | 1.38e-01 |
| rG_Ins_lg_and_S_cent_ins | Long insular gyrus and central sulcus of the insula | 1.19e-01 |
| rG_and_S_occipital_inf | Inferior occipital gyrus and sulcus | 4.15e-01 |
| rG_and_S_paracentral | Paracentral lobule and sulcus | 3.20e-01 |
| rG_oc-temp_med-Lingual | Lingual gyrus | 3.08e-01 |
| rG_temp_sup-Lateral | Lateral aspect of the superior temporal gyrus | 4.63e-01 |
| rLat_Fis-ant-Vertical | Vertical ramus of the anterior segment of the lateral sulcus | 2.10e-01 |
| rPole_temporal | Temporal pole | 2.64e-01 |
| rS_central | Central sulcus | 2.89e-01 |
| rS_circular_insula_ant | Anterior segment of the circular sulcus of the insula | 1.63e-01 |
| rS_circular_insula_inf | Inferior segment of the circular sulcus of the insula | 3.34e-01 |
| rS_circular_insula_sup | Superior segment of the circular sulcus of the insula | 3.45e-01 |
| rS_oc-temp_med_and_Lingual | Medial occipito-temporal sulcus and lingual sulcus | 4.84e-01 |
| rS_occipital_ant | Anterior occipital sulcus and preoccipital notch | 4.96e-01 |
| rS_orbital_lateral | Lateral orbital sulcus | 1.36e-01 |
| rS_pericallosal | Pericallosal sulcus | 1.28e-01 |
| rS_temporal_transverse | Transverse temporal sulcus | 2.24e-01 |
CAT12
| r_square | ||
|---|---|---|
| roiName | name | |
| lG_and_S_paracentral | Paracentral lobule and sulcus | 2.72e-01 |
| lG_cuneus | Cuneus | 2.41e-01 |
| lG_occipital_sup | Superior occipital gyrus | 5.13e-01 |
| lG_parietal_sup | Superior parietal lobule | 5.50e-01 |
| lG_precuneus | Precuneus | 5.06e-01 |
| lG_rectus | Straight gyrus | 1.35e-01 |
| lG_temp_sup-G_T_transv | Anterior transverse temporal gyrus | 3.11e-01 |
| lG_temp_sup-Lateral | Lateral aspect of the superior temporal gyrus | 4.32e-01 |
| lG_temp_sup-Plan_tempo | Planum temporale or temporal plane of the superior temporal gyrus | 5.82e-01 |
| lPole_occipital | Occipital pole | 2.97e-01 |
| lS_calcarine | Calcarine sulcus | 3.48e-01 |
| lS_central | Central sulcus | 3.41e-01 |
| lS_oc_sup_and_transversal | Superior occipital sulcus and transverse occipital sulcus | 5.12e-01 |
| lS_occipital_ant | Anterior occipital sulcus and preoccipital notch | 4.27e-01 |
| lS_precentral-inf-part | Inferior part of the precentral sulcus | 5.07e-01 |
| lS_temporal_inf | Inferior temporal sulcus | 2.33e-01 |
| lS_temporal_sup | Superior temporal sulcus | 5.80e-01 |
| lS_temporal_transverse | Transverse temporal sulcus | 4.51e-01 |
| rG_cuneus | Cuneus | 2.87e-01 |
| rG_front_sup | Superior frontal gyrus | 6.49e-01 |
| rG_occipital_middle | Middle occipital gyrus | 5.27e-01 |
| rG_occipital_sup | Superior occipital gyrus | 4.86e-01 |
| rG_precuneus | Precuneus | 5.60e-01 |
| rG_subcallosal | Subcallosal area | 5.11e-03 |
| rG_temp_sup-Plan_tempo | Planum temporale or temporal plane of the superior temporal gyrus | 5.97e-01 |
| rLat_Fis-post | Posterior ramus | 5.31e-01 |
| rS_calcarine | Calcarine sulcus | 4.16e-01 |
| rS_orbital_med-olfact | Medial orbital sulcus | 8.59e-02 |
| rS_precentral-inf-part | Inferior part of the precentral sulcus | 5.03e-01 |
df_cat12_important["type"] = "non-overlapping"
df_overlap["type"] = "overlapping"
df_fs_important["type"] = "non-overlapping"
df_fs_all = pd.concat([df_overlap, df_fs_important])
df_cat12_all = pd.concat([df_overlap, df_cat12_important])
fig = plt.figure(1, figsize=(10, 5))
ax = fig.add_subplot(1, 2, 1)
sns.stripplot(data=df_fs_all, x="type", y="r_square")
plt.xticks(rotation=20)
plt.ylabel("$R^2$")
plt.xlabel("")
plt.title("FreeSurfer")
ax = fig.add_subplot(1, 2, 2)
sns.stripplot(data=df_cat12_all, x="type", y="r_square")
plt.ylabel("$R^2$")
plt.xticks(rotation=20)
plt.xlabel("")
plt.title("CAT12")
plt.tight_layout()
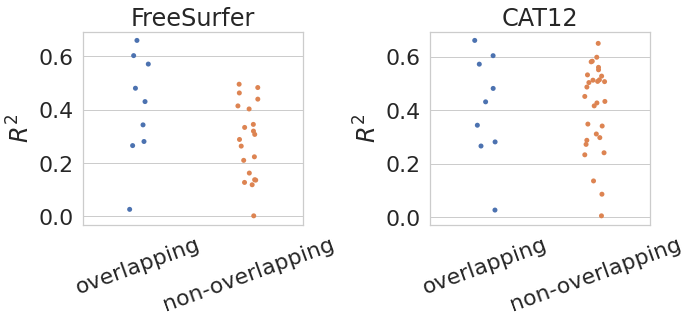
df_test_fs = pg.ttest(df_fs_all.loc[df_fs_all["type"] == "non-overlapping"]["r_square"],
df_fs_all.loc[df_fs_all["type"] == "overlapping"]["r_square"])
df_test_cat12 = pg.ttest(df_cat12_all.loc[df_cat12_all["type"] == "non-overlapping"]["r_square"],
df_cat12_all.loc[df_cat12_all["type"] == "overlapping"]["r_square"])
display(HTML(f"<br><h5>FreeSurfer</h5>"))
display(HTML(df_test_fs.to_html()))
display(HTML(f"<br><h5>CAT12</h5>"))
display(HTML(df_test_cat12.to_html()))
FreeSurfer
| T | dof | alternative | p-val | CI95% | cohen-d | BF10 | power | |
|---|---|---|---|---|---|---|---|---|
| T-test | -1.67e+00 | 1.17e+01 | two-sided | 1.22e-01 | [-0.28, 0.04] | 7.65e-01 | 0.997 | 4.51e-01 |
CAT12
| T | dof | alternative | p-val | CI95% | cohen-d | BF10 | power | |
|---|---|---|---|---|---|---|---|---|
| T-test | 3.79e-02 | 1.15e+01 | two-sided | 9.70e-01 | [-0.16, 0.16] | 1.61e-02 | 0.356 | 5.02e-02 |

