Results
Contents
Results¶
Data Loading¶
suppressWarnings(suppressMessages(library("tidyverse")))
suppressWarnings(suppressMessages(library("ggpubr")))
suppressWarnings(suppressMessages(library("rstatix")))
library(tidyverse)
library(ggpubr)
library(rstatix)
map_abb <- list("df_gm"= "DF+ǴM", "gm"= "GM")
all_results <- c()
df_eval <- list("crossval"= data.frame(), "test"= data.frame())
for (evaluation in names(df_eval)){
for (mod in names(map_abb)){
filename <- sprintf("%s_%s.csv", mod, evaluation)
path_col <- file.path(getwd(), "..", "..", "..", "resources", filename)
df_mod <- read.csv(path_col)
df_mod$fusion <- map_abb[[mod]]
df_eval[[evaluation]] <- rbind(df_eval[[evaluation]], df_mod)
}
}
df_val <- df_eval[["crossval"]]
df_test <- df_eval[["test"]]
Validation data¶
head(df_val)
| fold | r2 | MAE | fusion | |
|---|---|---|---|---|
| <int> | <dbl> | <dbl> | <chr> | |
| 1 | 0 | 0.8026467 | 5.740682 | DF+ǴM |
| 2 | 1 | 0.7473344 | 6.313860 | DF+ǴM |
| 3 | 2 | 0.7362875 | 5.625505 | DF+ǴM |
| 4 | 3 | 0.7456859 | 5.830985 | DF+ǴM |
| 5 | 4 | 0.7941903 | 5.643035 | DF+ǴM |
| 6 | 5 | 0.7565052 | 5.573205 | DF+ǴM |
aggregate(df_val[,c("r2", "MAE")], list(df_val$fusion), mean)
| Group.1 | r2 | MAE |
|---|---|---|
| <chr> | <dbl> | <dbl> |
| DF+ǴM | 0.7884606 | 5.548079 |
| GM | 0.7508428 | 6.140916 |
Test data¶
head(df_test)
| subjectID | gender | age | y_hat | fusion | |
|---|---|---|---|---|---|
| <chr> | <int> | <dbl> | <dbl> | <chr> | |
| 1 | sub-474 | 1 | 34.01 | 40.33901 | DF+ǴM |
| 2 | sub-292 | 1 | 23.73 | 31.65292 | DF+ǴM |
| 3 | sub-382 | 1 | 65.91 | 46.45294 | DF+ǴM |
| 4 | sub-434 | 1 | 67.24 | 56.62014 | DF+ǴM |
| 5 | sub-395 | 0 | 45.03 | 35.61988 | DF+ǴM |
| 6 | sub-306 | 1 | 30.18 | 39.87024 | DF+ǴM |
df_test["abs_diff"] <- abs(df_test$y_hat - df_test$age)
result_mean <- aggregate(df_test[,c("age", "y_hat", "abs_diff")], list(df_test$fusion), mean)
result_mean
| Group.1 | age | y_hat | abs_diff |
|---|---|---|---|
| <chr> | <dbl> | <dbl> | <dbl> |
| DF+ǴM | 42.37309 | 45.59776 | 6.896464 |
| GM | 42.37309 | 45.49686 | 7.956941 |
Statistics¶
Paired t-test¶
colors <- c("#AB1111", "#FFC107")
pwc <- df_val %>%
pairwise_t_test(
MAE ~ fusion, paired = TRUE, alternative="less"
)
pwc
options(repr.plot.width = 7, repr.plot.height = 5)
bxp <- ggboxplot(df_val, x = "fusion", y = "MAE", add = "point", fill = colors,
xlab = "Fusion", ylab = "MAE") + theme(text = element_text(size=20))
pwc <- pwc %>% add_xy_position(x = "fusion")
bxp <- bxp +
stat_pvalue_manual(pwc, label = "p = {p.adj}") +
labs(
subtitle = get_test_label(pwc, detailed = TRUE),
) + geom_point(data = data.frame(x = factor(result_mean[["Group.1"]]), y = result_mean[["abs_diff"]]),
aes(x=x, y=y),
color = 'red', shape=8, size=5)
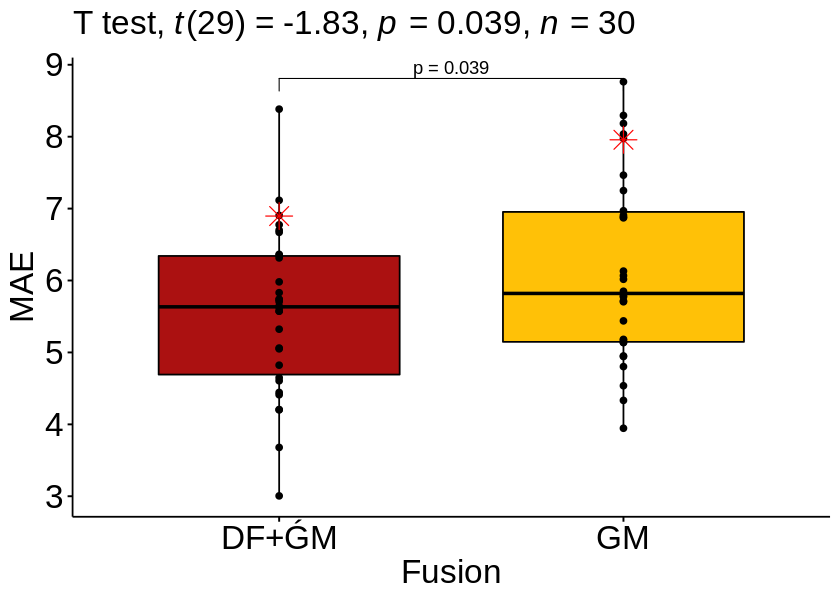
bxp
| .y. | group1 | group2 | n1 | n2 | statistic | df | p | p.adj | p.adj.signif | |
|---|---|---|---|---|---|---|---|---|---|---|
| <chr> | <chr> | <chr> | <int> | <int> | <dbl> | <dbl> | <dbl> | <dbl> | <chr> | |
| 1 | MAE | DF+ǴM | GM | 30 | 30 | -1.828452 | 29 | 0.039 | 0.039 | * |
Warning message in if (fill %in% names(data) & is.null(add.params$fill)) add.params$fill <- fill:
“the condition has length > 1 and only the first element will be used”