Results
Contents
Results¶
Data Loading¶
suppressWarnings(suppressMessages(library("tidyverse")))
suppressWarnings(suppressMessages(library("ggpubr")))
suppressWarnings(suppressMessages(library("rstatix")))
suppressWarnings(suppressMessages(library("plyr")))
library(tidyverse)
library(ggpubr)
library(rstatix)
library(plyr)
map_abb <- list("df"="DF", "gm"="GM","wm"="WM", "csf"="CSF")
df_eval <- list("crossval"= data.frame(), "test"= data.frame())
for (evaluation in names(df_eval)){
for (mod in names(map_abb)){
filename <- sprintf("%s_%s.csv", mod, evaluation)
path_col <- file.path(getwd(), "..", "..", "..", "resources", filename)
df_mod <- read.csv(path_col)
df_mod$modality <- map_abb[[mod]]
df_eval[[evaluation]] <- rbind(df_eval[[evaluation]], df_mod)
}
}
df_val <- df_eval[["crossval"]]
df_test <- df_eval[["test"]]
Validation data¶
head(df_val)
| fold | MAE | r2 | modality | |
|---|---|---|---|---|
| <int> | <dbl> | <dbl> | <chr> | |
| 1 | 0 | 4.910286 | 0.8573168 | DF |
| 2 | 1 | 4.177728 | 0.7937252 | DF |
| 3 | 2 | 6.336831 | 0.7525602 | DF |
| 4 | 3 | 6.220759 | 0.7729788 | DF |
| 5 | 4 | 5.494937 | 0.7465731 | DF |
| 6 | 5 | 6.471542 | 0.7765924 | DF |
aggregate(df_val[,c("r2", "MAE")], list(df_val$modality), mean)
| Group.1 | r2 | MAE |
|---|---|---|
| <chr> | <dbl> | <dbl> |
| CSF | 0.7068463 | 6.609508 |
| DF | 0.7616746 | 5.843714 |
| GM | 0.7508428 | 6.140916 |
| WM | 0.7235893 | 6.501720 |
Test data¶
head(df_test)
| subjectID | gender | age | y_hat | modality | |
|---|---|---|---|---|---|
| <chr> | <int> | <dbl> | <dbl> | <chr> | |
| 1 | sub-517 | 0 | 36.16 | 40.94362 | DF |
| 2 | sub-303 | 0 | 25.46 | 37.59454 | DF |
| 3 | sub-232 | 1 | 28.81 | 35.01436 | DF |
| 4 | sub-462 | 1 | 75.08 | 67.79715 | DF |
| 5 | sub-331 | 0 | 23.49 | 30.64490 | DF |
| 6 | sub-294 | 1 | 27.08 | 29.80392 | DF |
df_test["abs_diff"] <- abs(df_test$y_hat - df_test$age)
result_mean <- aggregate(df_test[,c("age", "y_hat", "abs_diff")], list(df_test$modality), mean)
result_mean
| Group.1 | age | y_hat | abs_diff |
|---|---|---|---|
| <chr> | <dbl> | <dbl> | <dbl> |
| CSF | 42.37309 | 49.54023 | 9.124851 |
| DF | 42.37309 | 46.25928 | 7.096847 |
| GM | 42.37309 | 45.49686 | 7.956941 |
| WM | 42.37309 | 37.31041 | 9.990654 |
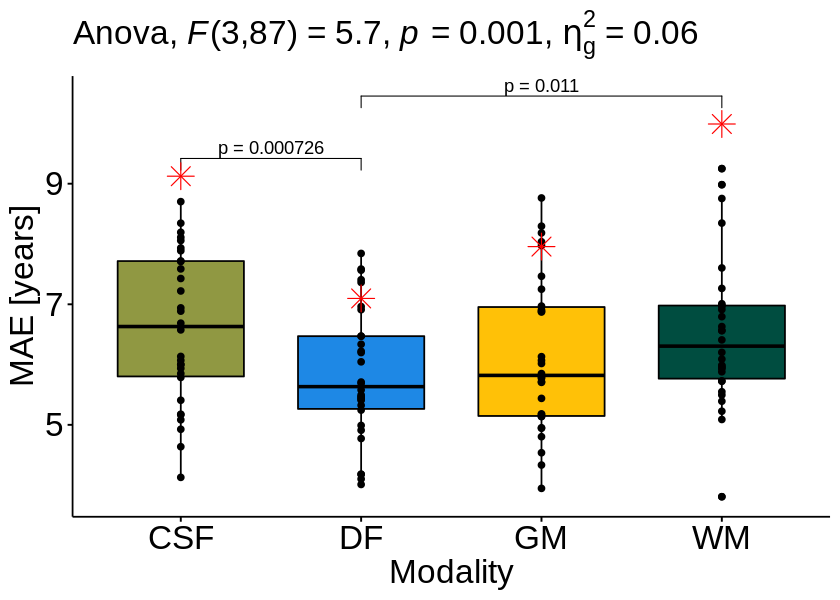
Statistics¶
Repeated Measures ANOVA¶
df_val$modality <- factor(df_val$modality)
res.aov <- anova_test(data = df_val, dv = MAE, wid = fold, within = modality)
get_anova_table(res.aov)
| Effect | DFn | DFd | F | p | p<.05 | ges | |
|---|---|---|---|---|---|---|---|
| <chr> | <dbl> | <dbl> | <dbl> | <dbl> | <chr> | <dbl> | |
| 1 | modality | 3 | 87 | 5.701 | 0.001 | * | 0.06 |
Post-hoc analysis¶
pwc <- df_val %>%
pairwise_t_test(
MAE ~ modality, paired = TRUE,
p.adjust.method = "bonferroni"
)
pwc <- pwc %>% add_xy_position(x = "modality")
pwc
| .y. | group1 | group2 | n1 | n2 | statistic | df | p | p.adj | p.adj.signif | y.position | groups | xmin | xmax |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <chr> | <chr> | <chr> | <int> | <int> | <dbl> | <dbl> | <dbl> | <dbl> | <chr> | <dbl> | <named list> | <dbl> | <dbl> |
| MAE | CSF | DF | 30 | 30 | 4.4361533 | 29 | 0.000121 | 0.000726 | *** | 9.418840 | CSF, DF | 1 | 2 |
| MAE | CSF | GM | 30 | 30 | 2.0552780 | 29 | 0.049000 | 0.294000 | ns | 9.621448 | CSF, GM | 1 | 3 |
| MAE | CSF | WM | 30 | 30 | 0.4207693 | 29 | 0.677000 | 1.000000 | ns | 9.824056 | CSF, WM | 1 | 4 |
| MAE | DF | GM | 30 | 30 | -1.5010999 | 29 | 0.144000 | 0.864000 | ns | 10.026664 | DF, GM | 2 | 3 |
| MAE | DF | WM | 30 | 30 | -3.4434066 | 29 | 0.002000 | 0.011000 | * | 10.229272 | DF, WM | 2 | 4 |
| MAE | GM | WM | 30 | 30 | -1.9403815 | 29 | 0.062000 | 0.373000 | ns | 10.431880 | GM, WM | 3 | 4 |
colors <- c("#909842", "#1E88E5", "#FFC107", "#004D40")
options(repr.plot.width = 7, repr.plot.height = 5)
bxp <- ggboxplot(df_val, x = "modality", y = "MAE", add = "point",
xlab = "Modality", ylab = "MAE [years]", fill=colors) + theme(text = element_text(size=20))
bxp <- bxp +
stat_pvalue_manual(pwc, hide.ns = TRUE, coord.flip=FALSE, label = "p = {p.adj}", step.increase=0.035) +
labs(
subtitle = get_test_label(res.aov, detailed = TRUE),
)
# add the MAE results for the test set
bxp <- bxp + geom_point(data = data.frame(x = factor(result_mean[["Group.1"]]), y = result_mean[["abs_diff"]]),
aes(x=x, y=y),
color = 'red', shape=8, size=5)
bxp
Warning message in if (fill %in% names(data) & is.null(add.params$fill)) add.params$fill <- fill:
“the condition has length > 1 and only the first element will be used”